2LCU
 
 | | NMR structure of BC28.1 | | Descriptor: | Bc28.1 | | Authors: | Roumestand, C, Delbecq, S, Yang, Y. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Bc28.1, Major Erythrocyte-binding Protein from Babesia canis Merozoite Surface.
J.Biol.Chem., 287, 2012
|
|
2LC0
 
 | | Rv0020c_Nter structure | | Descriptor: | Putative uncharacterized protein TB39.8 | | Authors: | Barthe, P.P, Cohen-Gonsaud, M.M, Roumestand, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Mycobacterium tuberculosis Rv0020c Protein and Its Interaction with the PknB Kinase
Structure, 19, 2011
|
|
9F5H
 
 | | Crystal structure of MGAT5 bump-and-hole mutant in complex with UDP and M592 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Secreted alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Liu, Y, Bineva-Todd, G, Meek, R, Mazo, L, Piniello, B, Moroz, O.V, Begum, N, Roustan, C, Tomita, S, Kjaer, S, Rovira, C, Davies, G.J, Schumann, B. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Bioorthogonal Precision Tool for Human N -Acetylglucosaminyltransferase V.
J.Am.Chem.Soc., 146, 2024
|
|
1CXN
 
 | | REFINED THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SNAKE CARDIOTOXIN: ANALYSIS OF THE SIDE-CHAIN ORGANISATION SUGGESTS THE EXISTENCE OF A POSSIBLE PHOSPHOLIPID BINDING SITE | | Descriptor: | CARDIOTOXIN GAMMA | | Authors: | Gilquin, B, Roumestand, C, Zinn-Justin, S, Menez, A, Toma, F. | | Deposit date: | 1994-07-08 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Refined three-dimensional solution structure of a snake cardiotoxin: analysis of the side-chain organization suggests the existence of a possible phospholipid binding site.
Biopolymers, 33, 1993
|
|
1CXO
 
 | | REFINED THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SNAKE CARDIOTOXIN: ANALYSIS OF THE SIDE-CHAIN ORGANISATION SUGGESTS THE EXISTENCE OF A POSSIBLE PHOSPHOLIPID BINDING SITE | | Descriptor: | CARDIOTOXIN GAMMA | | Authors: | Gilquin, B, Roumestand, C, Zinn-Justin, S, Menez, A, Toma, F. | | Deposit date: | 1994-11-07 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Refined three-dimensional solution structure of a snake cardiotoxin: analysis of the side-chain organization suggests the existence of a possible phospholipid binding site.
Biopolymers, 33, 1993
|
|
1D5Q
 
 | | SOLUTION STRUCTURE OF A MINI-PROTEIN REPRODUCING THE CORE OF THE CD4 SURFACE INTERACTING WITH THE HIV-1 ENVELOPE GLYCOPROTEIN | | Descriptor: | CHIMERIC MINI-PROTEIN | | Authors: | Vita, C, Drakopoulou, E, Vizzanova, J, Rochette, S, Martin, L, Menez, A, Roumestand, C, Yang, Y.S, Ylisastigui, L, Benjouad, A, Gluckman, J.C. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6ZUO
 
 | | Human RIO1(kd)-StHA late pre-40S particle, structural state A (pre 18S rRNA cleavage) | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
6ZV6
 
 | | Human RIO1(kd)-StHA late pre-40S particle, structural state B (post 18S rRNA cleavage) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
7B62
 
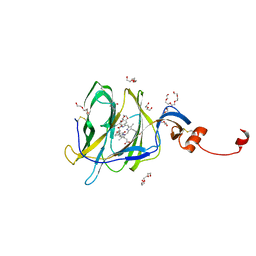 | | Crystal structure of SARS-CoV-2 spike protein N-terminal domain in complex with biliverdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pye, V.E, Rosa, A, Roustan, C, Cherepanov, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
6Y7C
 
 | | Early cytoplasmic yeast pre-40S particle (purified with Tsr1 as bait) | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Plassart, L, Plisson-Chastang, C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Good Vibrations: Structural Remodeling of Maturing Yeast Pre-40S Ribosomal Particles Followed by Cryo-Electron Microscopy.
Molecules, 25, 2020
|
|
7ZJY
 
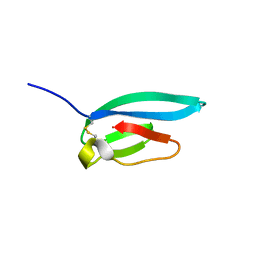 | | The NMR structure of the MAX67 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7ZK0
 
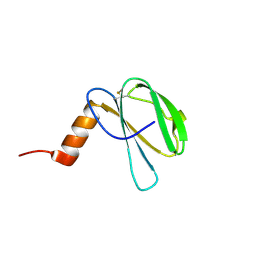 | | The NMR structure of the MAX60 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7ZKD
 
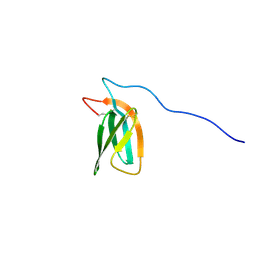 | | The NMR structure of the MAX47 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
1XSF
 
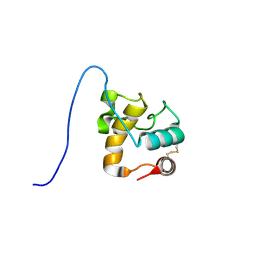 | | Solution structure of a resuscitation promoting factor domain from Mycobacterium tuberculosis | | Descriptor: | Probable resuscitation-promoting factor rpfB | | Authors: | Cohen-Gonsaud, M, Barthe, P, Henderson, B, Ward, J, Roumestand, C, Keep, N.H. | | Deposit date: | 2004-10-19 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes
Nat.Struct.Mol.Biol., 12, 2005
|
|
2CRD
 
 | | ANALYSIS OF SIDE-CHAIN ORGANIZATION ON A REFINED MODEL OF CHARYBDOTOXIN: STRUCTURAL AND FUNCTIONAL IMPLICATIONS | | Descriptor: | CHARYBDOTOXIN | | Authors: | Bontems, F, Roumestand, C, Gilquin, B, Menez, A, Toma, F. | | Deposit date: | 1993-02-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Analysis of side-chain organization on a refined model of charybdotoxin: structural and functional implications.
Biochemistry, 31, 1992
|
|
1BGK
 
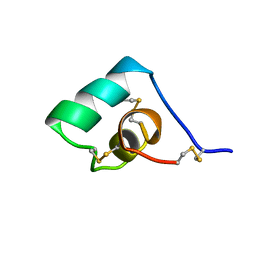 | | SEA ANEMONE TOXIN (BGK) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL, NMR, 15 STRUCTURES | | Descriptor: | BGK | | Authors: | Dauplais, M, Lecoq, A, Song, J, Cotton, J, Jamin, N, Gilquin, B, Roumestand, C, Vita, C, Harvey, A, Menez, A. | | Deposit date: | 1996-05-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures.
J.Biol.Chem., 272, 1997
|
|
8B7D
 
 | | Luminal domain of TMEM106B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Pye, V.E, Roustan, C, Cherepanov, P. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry.
Cell, 186, 2023
|
|
1SXM
 
 | | SCORPION TOXIN (NOXIUSTOXIN) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL AND LOW AFFINITY FOR CALCIUM DEPENDENT POTASSIUM CHANNEL (NMR AT 20 DEGREES, PH3.5, 39 STRUCTURES) | | Descriptor: | NOXIUSTOXIN | | Authors: | Dauplais, M, Gilquin, B, Possani, L.D, Gurrola-Briones, G, Roumestand, C, Menez, A. | | Deposit date: | 1995-09-07 | | Release date: | 1996-01-29 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional solution structure of noxiustoxin: analysis of structural differences with related short-chain scorpion toxins.
Biochemistry, 34, 1995
|
|
1YYL
 
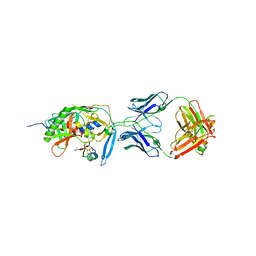 | | crystal structure of CD4M33, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD4M33, scorpion-toxin mimic of CD4, ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
1YYM
 
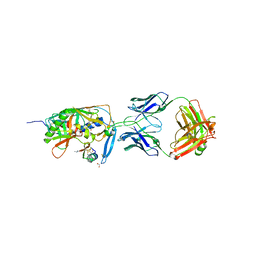 | | crystal structure of F23, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120), ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
2LUD
 
 | |
8C8A
 
 | | The NMR structure of the MAX28 effector from Magnaporthe oryzae | | Descriptor: | But2 domain-containing protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Kroj, T, Roumestand, C, Barthe, P. | | Deposit date: | 2023-01-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
2KB3
 
 | | NMR Structure of the phosphorylated form of OdhI, pOdhI. | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Barthe, P, Roumestand, C, Canova, M, Hurard, C, Molle, V, Cohen-Gonsaud, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Dynamic and Structural Characterization of a Bacterial FHA Protein Reveals a New Autoinhibition Mechanism.
Structure, 17, 2009
|
|
2KFS
 
 | | NMR structure of Rv2175c | | Descriptor: | Conserved hypothetical regulatory protein | | Authors: | Barthe, P, Cohen-Gonsaud, M, Roumestand, C, Molle, V. | | Deposit date: | 2009-02-27 | | Release date: | 2009-05-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The Mycobacterium tuberculosis Ser/Thr Kinase Substrate Rv2175c Is a DNA-binding Protein Regulated by Phosphorylation.
J.Biol.Chem., 284, 2009
|
|
7NRN
 
 | | NMR structure of GIPC1-GH2 domain | | Descriptor: | PDZ domain-containing protein GIPC1 | | Authors: | Barthe, P, Roumestand, C. | | Deposit date: | 2021-03-04 | | Release date: | 2021-04-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pressure and Chemical Unfolding of an alpha-Helical Bundle Protein: The GH2 Domain of the Protein Adaptor GIPC1.
Int J Mol Sci, 22, 2021
|
|
