2VIV
 
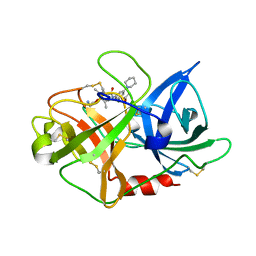 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-N-(3-chloro-5-piperidin-1-ylphenyl)-3,5-dimethylbenzamide, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator.
J.Med.Chem., 51, 2008
|
|
2VIQ
 
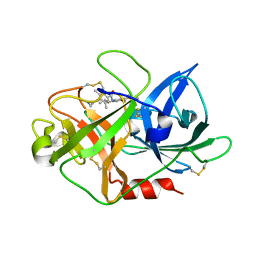 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-N-(2,5-diethoxyphenyl)-3,5-dimethylbenzamide, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator.
J.Med.Chem., 51, 2008
|
|
2VIO
 
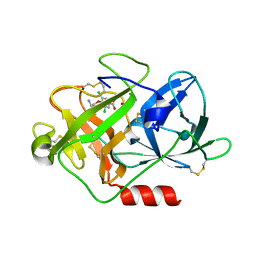 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-3,5-dichlorobenzoic acid, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator
J.Med.Chem., 51, 2008
|
|
2W6L
 
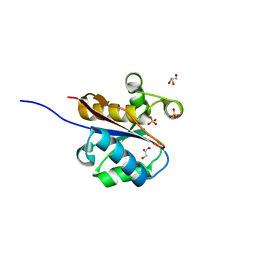 | | The crystal structure at 1.7 A resolution of CobE, a protein from the cobalamin (vitamin B12) biosynthetic pathway | | Descriptor: | COBE, GLYCEROL, SULFATE ION | | Authors: | Vevodova, J, Smith, D, McGoldrick, H, Deery, E, Murzin, A.G, Warren, M.J, Wilson, K.S. | | Deposit date: | 2008-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure at 1.7 A Resolution of Cobe, a Protein from the Cobalamin (Vitamin B12) Biosynthetic Pathway
To be Published
|
|
2W6K
 
 | | The crystal structure at 1.7 A resolution of CobE, a protein from the cobalamin (vitamin B12) biosynthetic pathway | | Descriptor: | COBE, GLYCEROL, SULFATE ION | | Authors: | Vevodova, J, Smith, D, McGoldrick, H, Deery, E, Murzin, A.G, Warren, M.J, Wilson, K.S. | | Deposit date: | 2008-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure at 1.7 A Resolution of Cobe, a Protein from the Cobalamin (Vitamin B12) Biosynthetic Pathway
To be Published
|
|
2I5G
 
 | | Crystal strcuture of amidohydrolase from Pseudomonas aeruginosa | | Descriptor: | amidohydrolase | | Authors: | Min, T, Sauder, J.M, Wasserman, S.R, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of amidohydrolase from Pseudomonas aeruginosa
To be Published
|
|
1QJV
 
 | | Pectin methylesterase PemA from Erwinia chrysanthemi | | Descriptor: | CHLORIDE ION, PECTIN METHYLESTERASE | | Authors: | Jenkins, J, Mayans, O, Smith, D, Worboys, K, Pickersgill, R. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Three-Dimensional Structure of Erwinia Chrysanthemi Pectin Methylesterase Reveals a Novel Esterase Active Site
J.Mol.Biol., 305, 2001
|
|
2IJR
 
 | | Crystal structure of a protein api92 from Yersinia pseudotuberculosis, Pfam DUF1281 | | Descriptor: | Hypothetical protein api92 | | Authors: | Jin, X, Min, T, Bonanno, J.B, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a hypothetical protein from Yersinia
pseudotuberculosis
To be Published
|
|
4B0Z
 
 | | Crystal structure of S. pombe Rpn12 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN12, GLYCEROL, MONOTHIOGLYCEROL, ... | | Authors: | Boehringer, J, Riedinger, C, Paraskevopoulos, K, Johnson, E.O.D, Lowe, E.D, Khoudian, C, Smith, D, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-12 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural and Functional Characterisation of Rpn12 Identifies Residues Required for Rpn10 Proteasome Incorporation.
Biochem.J., 448, 2012
|
|
2NPO
 
 | | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Acetyltransferase | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
2NLY
 
 | | Crystal structure of protein BH1492 from Bacillus halodurans, Pfam DUF610 | | Descriptor: | Divergent polysaccharide deacetylase hypothetical protein, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-20 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical protein BH1492 from Bacillus halodurans C-125
To be Published
|
|
2NRH
 
 | | Crystal structure of conserved putative Baf family transcriptional activator from Campylobacter jejuni | | Descriptor: | SULFATE ION, Transcriptional activator, putative, ... | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of conserved putative Baf family transcriptional activator from Campylobacter jejuni
To be Published
|
|
2OHW
 
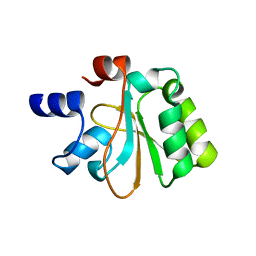 | | Crystal structure of the YueI protein from Bacillus subtilis | | Descriptor: | YueI protein | | Authors: | Bonanno, J.B, Jin, X, Mu, H, Dickey, M, Bain, K.T, Wu, B, Chen, T, Reyes, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-10 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the YueI protein from Bacillus subtilis
To be Published
|
|
2NR4
 
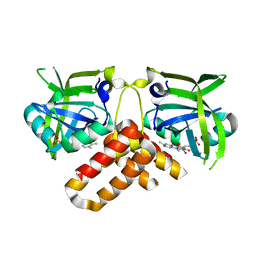 | | Crystal structure of FMN-bound protein MM1853 from Methanosarcina mazei, Pfam DUF447 | | Descriptor: | Conserved hypothetical protein, FLAVIN MONONUCLEOTIDE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Lau, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of conserved FMN bound hypothetical protein from Methanosarcina mazei
To be Published
|
|
2NWI
 
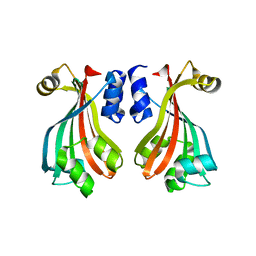 | | Crystal structure of protein AF1396 from Archaeoglobus fulgidus, Pfam DUF98 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Adams, J, Sridhar, V, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hypothetical protein O28875 from Archaeoglobus fulgidus
To be Published
|
|
2NS9
 
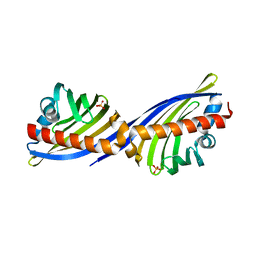 | | Crystal structure of protein APE2225 from Aeropyrum pernix K1, Pfam COXG | | Descriptor: | Hypothetical protein APE2225, PHOSPHATE ION | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein APE2225 from Aeropyrum pernix K1
To be Published
|
|
2OOG
 
 | | Crystal structure of glycerophosphoryl diester phosphodiesterase from Staphylococcus aureus | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SULFATE ION, ... | | Authors: | Patskovsky, Y, Fedorov, E, Toro, R, Sauder, J.M, Smith, D, Freeman, J, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase from Staphylococcus Aureus
To be Published
|
|
2OYN
 
 | | Crystal structure of CDP-bound protein MJ0056 from Methanococcus jannaschii, Pfam DUF120 | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, Hypothetical protein MJ0056, SODIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of hypothetical protein from Methanococcus jannaschii bound to CDP
TO BE PUBLISHED
|
|
2PCS
 
 | | Crystal structure of conserved protein from Geobacillus kaustophilus | | Descriptor: | Conserved protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Wu, B, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of conserved protein from Geobacillus kaustophilus
To be Published
|
|
2OZ8
 
 | | Crystal structure of putative mandelate racemase from Mesorhizobium loti | | Descriptor: | Mll7089 protein, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-25 | | Release date: | 2007-03-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of putative mandelate racemase from Mesorhizobium loti
To be Published
|
|
2PCE
 
 | | Crystal structure of putative mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM | | Descriptor: | PHOSPHATE ION, putative mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM
To be Published
|
|
2P61
 
 | | Crystal structure of protein TM1646 from Thermotoga maritima, Pfam DUF327 | | Descriptor: | Hypothetical protein TM_1646 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein TM_1646 from Thermotoga maritima
To be Published
|
|
2P1J
 
 | | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima | | Descriptor: | DNA polymerase III polC-type | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Izuka, M, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima
To be Published
|
|
2OX4
 
 | | Crystal structure of putative dehydratase from Zymomonas mobilis ZM4 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Dehydratase from Zymomonas Mobilis Zm4
To be Published
|
|
2OZ3
 
 | | Crystal structure of L-Rhamnonate dehydratase from Azotobacter vinelandii | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wu, B, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Rhamnonate dehydratase from azotobacter vinelandii
To be Published
|
|
