3NIU
 
 | | Crystal structure of the complex of dimeric goat lactoperoxidase with diethylene glycol at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vikram, G, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of the complex of dimeric goat lactoperoxidase with diethylene glycol at 2.9 A resolution
To be Published
|
|
3N3X
 
 | | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Vikram, G, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution
To be Published
|
|
3NFM
 
 | | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution
To be Published
|
|
3NJS
 
 | | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution
To be Published
|
|
3NAK
 
 | | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite at 3.3 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vikram, G, Singh, R.P, Singh, A.K, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite at 3.3 A resolution
To be Published
|
|
3NJU
 
 | | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Kaushik, S, Prem Kumar, R, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution
To be Published
|
|
3NX9
 
 | | Crystal structure of type I ribosome inactivating protein in complex with maltose at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-13 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of type I ribosome inactivating protein in complex with maltose at 1.7A resolution
To be Published
|
|
3O97
 
 | | Crystal Structure of the complex of C-lobe of lactoferrin with indole acetic acid at 2.68 A Resolution | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Sinha, M, Bhushan, A, Vikram, G, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of the complex of C-lobe of lactoferrin with indole acetic acid at 2.68 A Resolution
To be Published
|
|
3O9N
 
 | | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Singh, A, Kumar, S, Sinha, M, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution
To be Published
|
|
3O4K
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) and lipoteichoic acid at 2.1 A resolution | | Descriptor: | (2S)-1-({3-O-[2-(acetylamino)-4-amino-2,4,6-trideoxy-beta-D-galactopyranosyl]-alpha-D-glucopyranosyl}oxy)-3-(heptanoyloxy)propan-2-yl (7Z)-pentadec-7-enoate, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of recognition of pathogen-associated molecular patterns and inhibition of proinflammatory cytokines by camel peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3NNO
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1, alpha-L-rhamnopyranose | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution
To be Published
|
|
3OGX
 
 | | Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of heparin binding to camel peptidoglycan recognition protein-S
Int J Biochem Mol Biol, 3, 2012
|
|
3OSZ
 
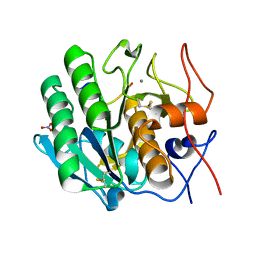 | | Crystal Structure of the complex of proteinase K with an antimicrobial nonapeptide, at 2.26 A resolution | | Descriptor: | 10-mer peptide, CALCIUM ION, NITRATE ION, ... | | Authors: | Singh, A, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-09-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of the complex of proteinase K with an antimicrobial nonapeptide, at 2.26 A resolution
To be Published
|
|
3OSH
 
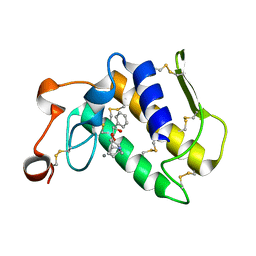 | | Crystal Structure of The Complex of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Shukla, P.K, Kaushik, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-09-09 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of The Complex of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution
To be Published
|
|
3P2J
 
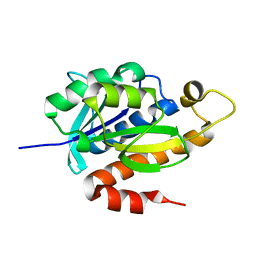 | | Crystal structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Yadav, R, Sinha, M, Arora, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-10-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution
To be Published
|
|
3PY4
 
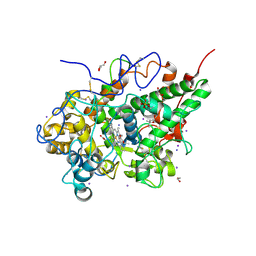 | | Crystal structure of bovine lactoperoxidase in complex with paracetamol at 2.4A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Sing, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-11 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of bovine lactoperoxidase in complex with paracetamol at 2.4A resolution
To be published
|
|
3PTL
 
 | | Crystal structure of proteinase K inhibited by a lactoferrin nonapeptide, Lys-Gly-Glu-Ala-Asp-Ala-Leu-Ser-Leu-Asp at 1.3 A resolution. | | Descriptor: | 10-mer peptide from Lactoferrin, Proteinase K | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of proteinase K inhibited by a lactoferrin nonapeptide, Lys-Gly-Glu-Ala-Asp-Ala-Leu-Ser-Leu-Asp at 1.3 A resolution.
TO BE PUBLISHED
|
|
3Q4Y
 
 | | Crystal structure of group I phospholipase A2 at 2.3 A resolution in 40% ethanol revealed the critical elements of hydrophobicity of the substrate-binding site | | Descriptor: | CALCIUM ION, ETHANOL, Phospholipase A2 isoform 3 | | Authors: | Shukla, P.K, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-26 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of group I phospholipase A2 at 2.3 A resolution in 40% ethanol revealed the critical elements of hydrophobicity of the substrate-binding site
To be Published
|
|
3Q4P
 
 | | Crystal structure of the complex of type I ribosome inactivating protein with 7n-methyl -8-hydroguanosine-5-p-diphosphate at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kushwaha, G.S, Yamini, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-24 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with 7n-methyl-8-hydroguanosine-5-p-diphosphate at 1.8 A resolution
To be Published
|
|
3QS0
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-02-19 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel AT 2.5 A resolution
To be Published
|
|
3QF1
 
 | | Crystal structure of the complex of caprine lactoperoxidase with diethylenediamine at 2.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-01-21 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of caprine lactoperoxidase with diethylenediamine at 2.6A resolution
To be Published
|
|
3QJ1
 
 | | Crystal structure of camel peptidoglycan recognition protein, PGRP-S with a trapped diethylene glycol in the ligand diffusion channel at 3.2 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Yamini, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-01-28 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of camel peptidoglycan recognition protein, PGRP-S with a trapped diethylene glycol in the ligand diffusion channel at 3.2 A resolution
To be Published
|
|
3QV4
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, ... | | Authors: | Shukla, P.K, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution
To be Published
|
|
3RT4
 
 | | Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Sharma, P, Dube, D, Singh, A, Mishra, B, Singh, N, Sinha, M, Dey, S, Kaur, P, Mitra, D.K, Sharma, S, Singh, T.P. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein.
J.Biol.Chem., 286, 2011
|
|
3RKE
 
 | | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution | | Descriptor: | (4-aminophenyl)-imidazol-1-yl-methanone, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dube, D, Singh, R.P, Sinha, M, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution
TO BE PUBLISHED
|
|
