7YOK
 
 | | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A | | Descriptor: | Cysteine synthase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A
To Be Published
|
|
7YOD
 
 | |
7YOH
 
 | |
7YOL
 
 | | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide of serine acetyltransferase from Haemophilus influenzae at 2.4 A | | Descriptor: | Cysteine synthase, Peptide from Serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Haemophilus influenzae at 2.4 A
To Be Published
|
|
7YOI
 
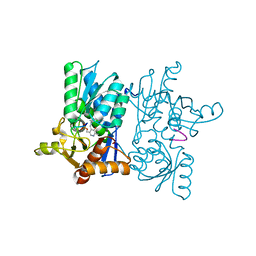 | |
7YOM
 
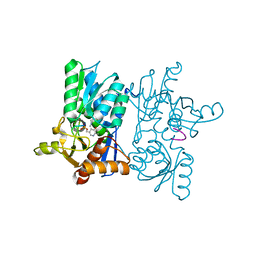 | | Crystal structure of tetra mutant (D67E,A68P,L98I,A301S) of O-acetylserine sulfhydrylase from Salmonella typhimurium in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.8 A | | Descriptor: | Cysteine synthase, peptide from serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of I88L single mutant of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.14 A
To Be Published
|
|
7YOE
 
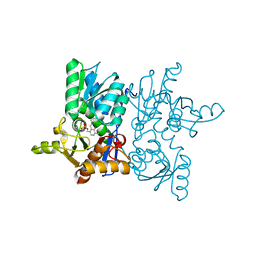 | |
5IWP
 
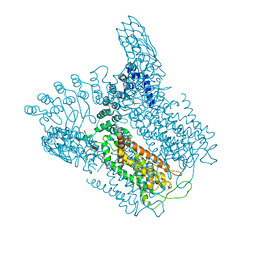 | | Structure of Transient Receptor Potential (TRP) channel TRPV6 in the presence of calcium | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Saotome, K, Singh, A.K, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structure of the epithelial calcium channel TRPV6.
Nature, 534, 2016
|
|
7OTK
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-233 | | Descriptor: | (2~{R})-2-[2-(6-aminopurin-9-yl)ethylamino]-3-phosphono-propanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTN
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-247 | | Descriptor: | (S)-2-((2-(6-amino-9H-purin-9-yl)ethyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OT6
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND inhibitor RMC-282 | | Descriptor: | (R)-N-(1-(6-amino-9H-purin-9-yl)propan-2-yl)-N-(2-phosphonoethyl)glycine, DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTA
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-230 | | Descriptor: | ((2-(6-amino-9H-purin-9-yl)ethyl)-L-seryl)phosphoramidic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
3PY4
 
 | | Crystal structure of bovine lactoperoxidase in complex with paracetamol at 2.4A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Sing, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-11 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of bovine lactoperoxidase in complex with paracetamol at 2.4A resolution
To be published
|
|
4MTV
 
 | | Crystal structure of the complex of Buffalo Signalling Glycoprotein with pentasaccharide at 2.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Shukla, P.K, Chaudhary, A, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the complex of Buffalo Signalling Glycoprotein with pentasaccharide at 2.8A resolution
To be Published
|
|
5DBH
 
 | |
6FQ4
 
 | | Structure of Chlamydial virulence factor TarP and vinculin head domain | | Descriptor: | TarP-VBS1, Vinculin | | Authors: | Whitewood, A.J, Singh, A.K, Brown, D.G, Goult, B.T. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chlamydial virulence factor TarP mimics talin to disrupt the talin-vinculin complex.
FEBS Lett., 592, 2018
|
|
4GWO
 
 | |
4GXA
 
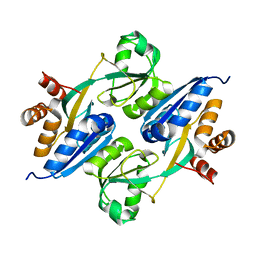 | |
4HO1
 
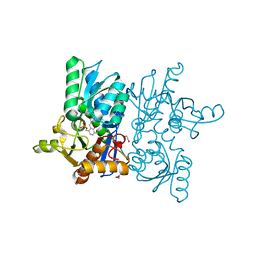 | |
4M4G
 
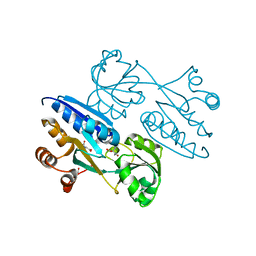 | | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, N-acetylserine. | | Descriptor: | DI(HYDROXYETHYL)ETHER, HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Mittal, M, Singh, A.K, Kumaran, S. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, N-acetylserine
To be Published
|
|
4LON
 
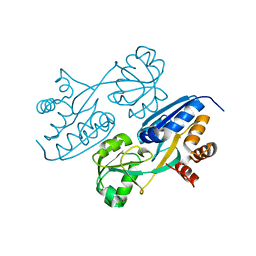 | |
4LQ5
 
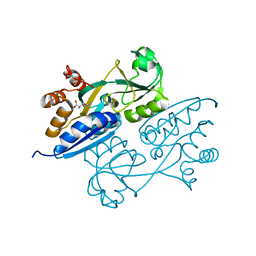 | | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, O-acetylserine at 2.8A | | Descriptor: | HTH-type transcriptional regulator CysB, O-ACETYLSERINE | | Authors: | Mittal, M, Singh, A.K, Kumaran, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | rystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, O-acetylserine at 2.8A
TO BE PUBLISHED
|
|
4LP2
 
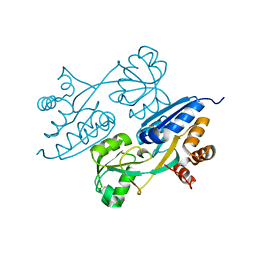 | |
4LQ2
 
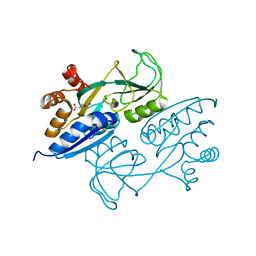 | | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium in complex with effector ligand, O-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, O-ACETYLSERINE | | Authors: | Mittal, M, Singh, A.K, Kumaran, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium in complex with effector ligand, O-acetylserine
TO BE PUBLISHED
|
|
4NU8
 
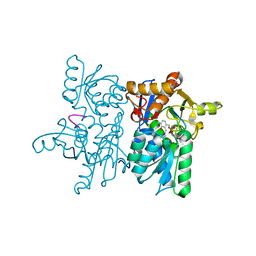 | |
