8BRW
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C | | Descriptor: | Methionine--tRNA ligase, ZINC ION | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRX
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C complexed with beta-3-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRV
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13M,I297C complexed with beta3-methionine. | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRU
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13M,I297C | | Descriptor: | CITRIC ACID, Methionine--tRNA ligase, ZINC ION | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
2O7O
 
 | | Crystal structure analysis of TetR(D) complex with doxycycline | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Aleksandrov, A, Proft, J, Hinrichs, W. | | Deposit date: | 2006-12-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Protonation Patterns in Tetracycline:Tet Repressor Recognition: Simulations and Experiments
Chembiochem, 8, 2007
|
|
1UL9
 
 | | CGL2 ligandfree | | Descriptor: | galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULC
 
 | | CGL2 in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULF
 
 | | CGL2 in complex with Blood Group A tetrasaccharide | | Descriptor: | alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULE
 
 | | CGL2 in complex with linear B2 trisaccharide | | Descriptor: | alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULD
 
 | | CGL2 in complex with blood group H type II | | Descriptor: | alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULG
 
 | | CGL2 in complex with Thomsen-Friedenreich antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
2WQ5
 
 | | Non-antibiotic properties of tetracyclines: structural basis for inhibition of secretory phospholipase A2. | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, CALCIUM ION, PHOSPHOLIPASE A2, ... | | Authors: | Dalm, D, Palm, G.J, Hinrichs, W. | | Deposit date: | 2009-08-13 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Non-Antibiotic Properties of Tetracyclines: Structural Basis for Inhibition of Secretory Phospholipase A(2).
J.Mol.Biol., 398, 2010
|
|
1FL1
 
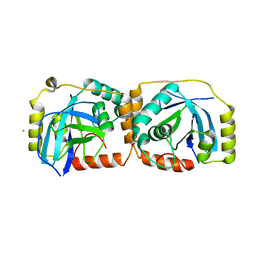 | | KSHV PROTEASE | | Descriptor: | POTASSIUM ION, PROTEASE | | Authors: | Reiling, K.K, Pray, T.R, Craik, C.S, Stroud, R.M. | | Deposit date: | 2000-08-11 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements.
Biochemistry, 39, 2000
|
|
2VPR
 
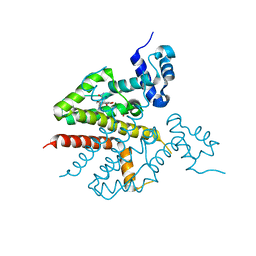 | | Tet repressor class H in complex with 5a,6- anhydrotetracycline-Mg | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Schuldt, L, Palm, G, Hinrichs, W. | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Tet Repressor Induction by Tetracycline: A Molecular Dynamics, Continuum Electrostatics, and Crystallographic Study
J.Mol.Biol., 378, 2008
|
|
1F5J
 
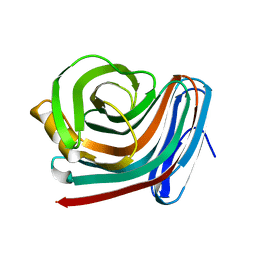 | | CRYSTAL STRUCTURE OF XYNB, A HIGHLY THERMOSTABLE BETA-1,4-XYLANASE FROM DICTYOGLOMUS THERMOPHILUM RT46B.1, AT 1.8 A RESOLUTION | | Descriptor: | BETA-1,4-XYLANASE, SULFATE ION | | Authors: | McCarthy, A.A, Baker, E.N. | | Deposit date: | 2000-07-26 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of XynB, a highly thermostable beta-1,4-xylanase from Dictyoglomus thermophilum Rt46B.1, at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1T99
 
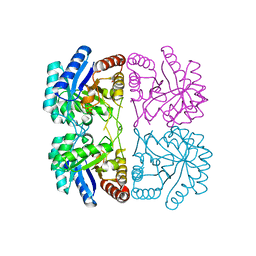 | | r106g kdo8ps without substrates | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, PHOSPHATE ION | | Authors: | Gatti, D.L. | | Deposit date: | 2004-05-16 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Effects of the Arg106==>Gly mutation on the catalytic and conformational cycle of Aquifex aeolicus KDO8P synthase.
To be Published
|
|
1T96
 
 | | r106g kdo8ps with pep | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Gatti, D.L. | | Deposit date: | 2004-05-14 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Effects of the Arg106==>Gly mutation on the catalytic and conformational cycle of Aquifex aeolicus KDO8P synthase.
To be Published
|
|
1T8X
 
 | | r106g kdo8ps with pep and a5p | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, CADMIUM ION, ... | | Authors: | Gatti, D.L. | | Deposit date: | 2004-05-13 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of the Arg106==>Gly mutation on the catalytic and conformational cycle of Aquifex aeolicus KDO8P synthase.
To be Published
|
|
1WS1
 
 | |
1OWM
 
 | | DATA1:DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1N51
 
 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWL
 
 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWO
 
 | | DATA4:photoreduced DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWP
 
 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
