4E0G
 
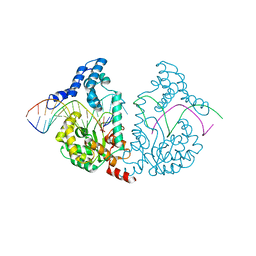 | | Protelomerase tela/DNA hairpin product/vanadate complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), Protelomerase, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2012-03-03 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
4E0Z
 
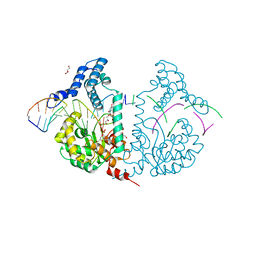 | | Protelomerase tela R205A covalently complexed with substrate DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*CP*A*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), GLYCEROL, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
4E0P
 
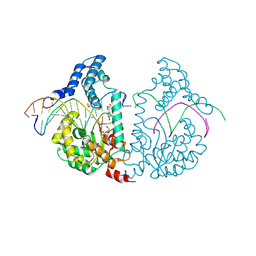 | | Protelomerase tela covalently complexed with substrate DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
4E0J
 
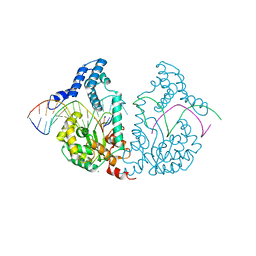 | | Protelomerase tela R255A mutant complexed with DNA hairpin product | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2012-03-04 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
4E0Y
 
 | | Protelomerase tela covalently complexed with mutated substrate DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*AP*T)-3'), DNA (5'-D(*CP*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), GLYCEROL, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
3EA6
 
 | | Atomic resolution of crystal structure of SEK | | Descriptor: | IODIDE ION, Staphylococcal enterotoxin K, ZINC ION | | Authors: | Shi, K, Huseby, M, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2008-08-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Structural Studies of an Emerging Pyrogenic Superantigen, SEK
To be Published
|
|
4FW1
 
 | |
4FW2
 
 | |
3SG8
 
 | |
3SGC
 
 | |
3SG9
 
 | |
6BUP
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica complexed with cyanuric acid | | Descriptor: | 1,3,5-triazine-2,4,6-triol, 1,3-PROPANDIOL, CALCIUM ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2017-12-11 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
6BUR
 
 | |
6BUN
 
 | |
6BUQ
 
 | |
6BUO
 
 | |
6BUM
 
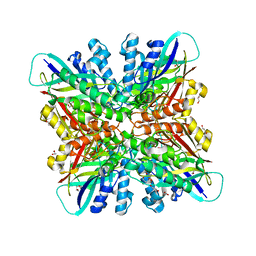 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica | | Descriptor: | 1,3-PROPANDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shi, K, Cho, S, Seffernick, J.L, Bera, A, Wackett, L.P, Aihara, H. | | Deposit date: | 2017-12-11 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
6NFK
 
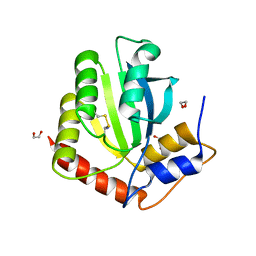 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
2G91
 
 | |
1DVL
 
 | |
1JO2
 
 | |
6NFM
 
 | |
1R3Z
 
 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*(5CM)P*GP*(5CM)P*GP*CP*GP*C)-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R41
 
 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*CP*GP*CP*GP*(5CM)P*GP*(5CM))-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1LAV
 
 | |
