1GFA
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFK
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFU
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GAY
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | MUTANT LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GF9
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFJ
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFG
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFR
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1CQ8
 
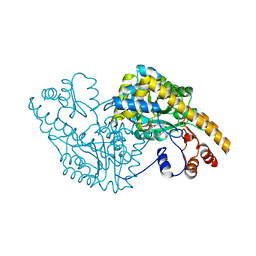 | | ASPARTATE AMINOTRANSFERASE (E.C. 2.6.1.1) COMPLEXED WITH C6-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-HEXANOIC ACID, ASPARTATE AMINOTRANSFERASE (2.6.1.1) | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1CQ7
 
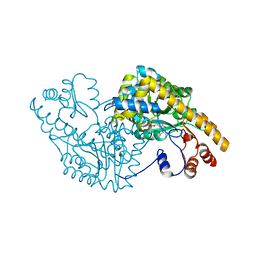 | | ASPARTATE AMINOTRANSFERASE (E.C. 2.6.1.1) COMPLEXED WITH C5-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-PENTANOIC ACID, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1CQ6
 
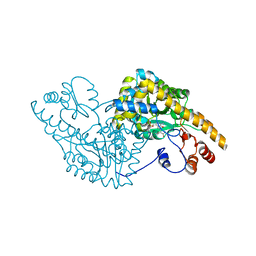 | | ASPARTATE AMINOTRANSFERASE COMPLEX WITH C4-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO- BUTYRIC ACID, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1K61
 
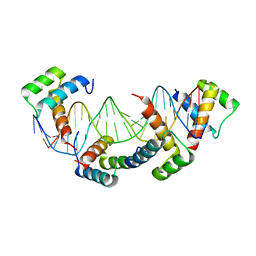 | | MATALPHA2 HOMEODOMAIN BOUND TO DNA | | Descriptor: | 5'-D(*(5IU)P*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*AP*CP*AP*TP*G)-3', 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*CP*AP*CP*GP*C)-3', Mating-type protein alpha-2 | | Authors: | Aishima, J, Gitti, R.K, Noah, J.E, Gan, H.H, Schlick, T, Wolberger, C. | | Deposit date: | 2001-10-14 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Hoogsteen base pair embedded in undistorted B-DNA
NUCLEIC ACIDS RES., 30, 2002
|
|
1L3X
 
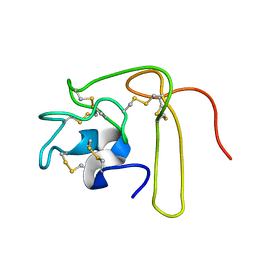 | | Solution Structure of Novel Disintegrin Salmosin | | Descriptor: | platelet aggregation inhibitor disintegrin | | Authors: | Shin, J, Lee, W. | | Deposit date: | 2002-03-01 | | Release date: | 2003-12-23 | | Last modified: | 2012-11-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel disintegrin, salmosin, from Agkistrondon halys venom
Biochemistry, 42, 2003
|
|
1EJQ
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE | | Descriptor: | SYNDECAN-4 | | Authors: | Shin, J, Oh, E.S, Lee, D, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-04 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE
To be Published
|
|
2LAV
 
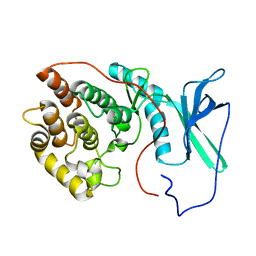 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
2LJP
 
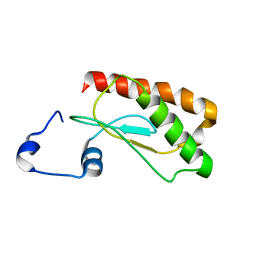 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for E.coli Ribonuclease P protein | | Descriptor: | Ribonuclease P protein component | | Authors: | Shin, J, Kim, K, Ryu, K, Han, K, Lee, Y, Choi, B. | | Deposit date: | 2011-09-21 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Escherichia coli C5 protein
To be Published
|
|
2N5Q
 
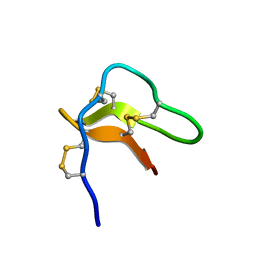 | | Solution structure of cystein-rich peptide jS1 from Jasminum sambac | | Descriptor: | cysteine-rich peptide jS1 | | Authors: | Shin, J, Kumari, G, Serra, A, Nguyen, P.Q.T, Yoon, H, Tam, J.P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification of a cysteine-rich peptide family with unusual disulfide connectivity from Jasminum sambac
To be Published
|
|
3W3X
 
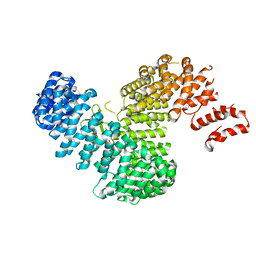 | | Crystal structure of Kap121p bound to Pho4p | | Descriptor: | Importin subunit beta-3, Phosphate system positive regulatory protein PHO4 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3Y
 
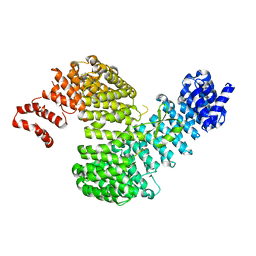 | | Crystal structure of Kap121p bound to Nup53p | | Descriptor: | Importin subunit beta-3, Nucleoporin NUP53 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3T
 
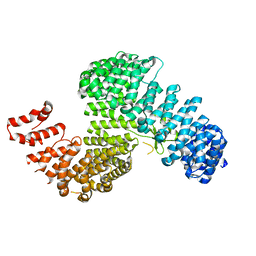 | | Crystal structure of Kap121p | | Descriptor: | Importin subunit beta-3 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3V
 
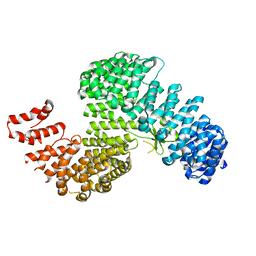 | |
1V4P
 
 | |
7VAX
 
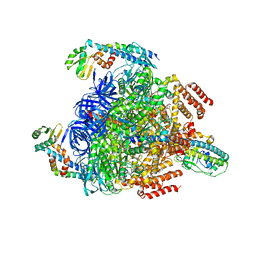 | | V1EG of V/A-ATPase from Thermus thermophilus at saturated ATP-gamma-S condition, state1-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAI
 
 | | V1EG of V/A-ATPase from Thermus thermophilus, state1-1 | | Descriptor: | V-type ATP synthase alpha chain, V-type ATP synthase beta chain, V-type ATP synthase subunit D, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAL
 
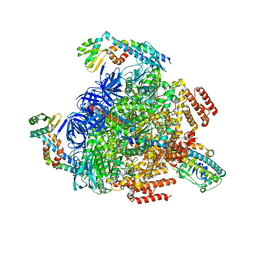 | | V1EG of V/A-ATPase from Thermus thermophilus, high ATP, state1-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
