6ZZF
 
 | |
7ACB
 
 | |
2JNI
 
 | | Spatial structure of antimicrobial peptide arenicin-2 in aqueous solution | | Descriptor: | Arenicin-2 | | Authors: | Ovchinnikova, T.V, Shenkarev, Z.O, Nadezhdin, K.D, Balandin, S.V, Zhmak, M.N, Kudelina, I.A, Finkina, E.I, Kokryakov, V.N, Arseniev, A.S. | | Deposit date: | 2007-01-25 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Recombinant expression, synthesis, purification, and solution structure of arenicin
Biochem.Biophys.Res.Commun., 360, 2007
|
|
2L03
 
 | |
2MJ0
 
 | |
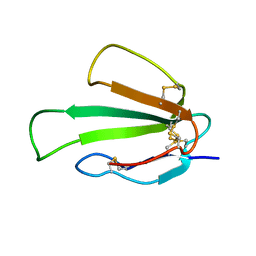
2MAL
 
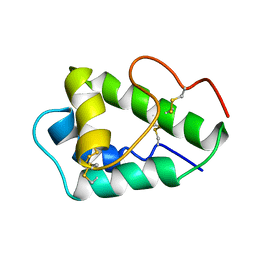 | |
2MQU
 
 | | Spatial structure of Hm-3, a membrane-active spider toxin affecting sodium channels | | Descriptor: | Neurotoxin Hm-3 | | Authors: | Myshkin, M.Y, Shenkarev, Z.O, Paramonov, A.S, Berkut, A.A, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2014-06-27 | | Release date: | 2014-11-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of membrane-active toxin from crab spider Heriaeus melloteei suggests parallel evolution of sodium channel gating modifiers in Araneomorphae and Mygalomorphae.
J. Biol. Chem., 290, 2015
|
|
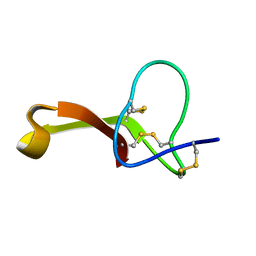
2N81
 
 | | Solution Structure of Lipid Transfer Protein From Pea Pisum Sativum | | Descriptor: | Lipid Transfer Protein | | Authors: | Paramonov, A.S, Rumynskiy, E.I, Bogdanov, I.V, Finkina, E.I, Melnikova, D.N, Ovchinnikova, T.V, Shenkarev, Z.O, Arseniev, A.S. | | Deposit date: | 2015-09-30 | | Release date: | 2016-05-11 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | A novel lipid transfer protein from the pea Pisum sativum: isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties.
BMC Plant Biol, 16
|
|
2N99
 
 | | Solution structure of the SLURP-2, a secreted isoform of Lynx1 | | Descriptor: | Ly-6/neurotoxin-like protein 1 | | Authors: | Paramonov, A.S, Shenkarev, Z.O, Lyukmanova, E.N, Arseniev, A.S. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Secreted Isoform of Human Lynx1 (SLURP-2): Spatial Structure and Pharmacology of Interactions with Different Types of Acetylcholine Receptors.
Sci Rep, 6, 2016
|
|
2LG4
 
 | |
2LJ7
 
 | |
1G2G
 
 | | MINIMAL CONFORMATION OF THE ALPHA-CONOTOXIN IMI FOR THE ALPHA7 NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR RECOGNITION | | Descriptor: | ALPHA-CONOTOXIN IMI | | Authors: | Lamthanh, H, Jegou-Matheron, C, Servent, D, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Minimal conformation of the alpha-conotoxin ImI for the alpha7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies.
FEBS Lett., 454, 1999
|
|
5LUE
 
 | | Minor form of the recombinant cytotoxin-1 from N. oxiana | | Descriptor: | VC-1=CYTOTOXIN | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
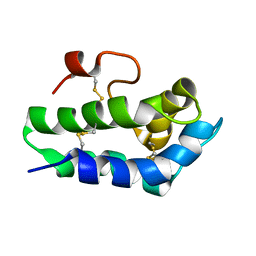
2MOA
 
 | |
