4YUU
 
 | | Crystal structure of oxygen-evolving photosystem II from a red alga | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ago, H, Shen, J.-R. | | Deposit date: | 2015-03-19 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7700038 Å) | | Cite: | Novel Features of Eukaryotic Photosystem II Revealed by Its Crystal Structure Analysis from a Red Alga
J.Biol.Chem., 291, 2016
|
|
1IZL
 
 | | Crystal Structure of Photosystem II | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Kamiya, N, Shen, J.-R. | | Deposit date: | 2002-10-04 | | Release date: | 2003-01-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4LJI
 
 | | Crystal structure at 1.5 angstrom resolution of the PsbV2 cytochrome from the cyanobacterium thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, Cytochrome c-550-like protein, HEME C | | Authors: | Suga, M, Lai, T.-L, Sugiura, M, Shen, J.-R, Boussac, A. | | Deposit date: | 2013-07-04 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Crystal structure at 1.5 angstrom resolution of the PsbV2 cytochrome from the cyanobacterium Thermosynechococcus elongatus
Febs Lett., 587, 2013
|
|
3A0B
 
 | | Crystal structure of Br-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3A0H
 
 | | Crystal structure of I-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1VCR
 
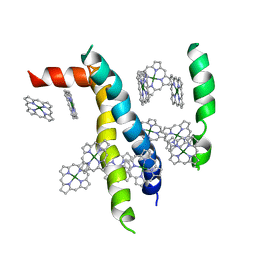 | | An icosahedral assembly of light-harvesting chlorophyll a/b protein complex from pea thylakoid membranes | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL B, Chlorophyll a-b binding protein AB80 | | Authors: | Hino, T, Kanamori, E, Shen, J.-R, Kouyama, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9.5 Å) | | Cite: | An icosahedral assembly of the light-harvesting chlorophyll a/b protein complex from pea chloroplast thylakoid membranes.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3WU2
 
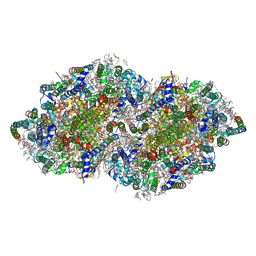 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Umena, Y, Kawakami, K, Shen, J.R, Kamiya, N. | | Deposit date: | 2014-04-21 | | Release date: | 2014-09-03 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A
Nature, 473, 2011
|
|
5V2C
 
 | | RE-REFINEMENT OF CRYSTAL STRUCTURE OF PHOTOSYSTEM II COMPLEX | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, J, Wiwczar, J.M, Brudvig, G.W. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chlorophyll a with a farnesyl tail in thermophilic cyanobacteria.
Photosyn. Res., 134, 2017
|
|
5Y5S
 
 | | Structure of photosynthetic LH1-RC super-complex at 1.9 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Yu, L.-J, Suga, M, Wang-Otomo, Z.-Y, Shen, J.-R. | | Deposit date: | 2017-08-09 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of photosynthetic LH1-RC supercomplex at 1.9 angstrom resolution.
Nature, 556, 2018
|
|
5ZE8
 
 | | Crystal structure of a penta-heme cytochrome c552 from Thermochromatium tepidum | | Descriptor: | GLYCEROL, HEME C, SULFATE ION, ... | | Authors: | Yu, L.-J, Chen, J.-H, Shen, J.-R. | | Deposit date: | 2018-02-27 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Properties and structure of a low-potential, penta-heme cytochrome c552from a thermophilic purple sulfur photosynthetic bacterium Thermochromatium tepidum.
Photosyn. Res., 139, 2019
|
|
4QS7
 
 | |
4QS9
 
 | |
4QS8
 
 | |
8WCK
 
 | | FCP tetramer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, Chlorophyll a/b-binding protein, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Shen, J.-R, Liu, C, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 5, 2024
|
|
8WCL
 
 | | FCP pentamer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Liu, C, Shen, J.-R, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 5, 2024
|
|
1NZE
 
 | | Crystal structure of PsbQ polypeptide of photosystem II from higher plants | | Descriptor: | Oxygen-evolving enhancer protein 3, ZINC ION | | Authors: | Calderone, V, Trabucco, M, Vujicic, A, Battistutta, R, Giacometti, G.M, Andreucci, F, Barbato, R, Zanotti, G. | | Deposit date: | 2003-02-17 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the PsbQ protein of photosystem II from higher plants
Embo Rep., 4, 2003
|
|
8GN2
 
 | | Crystal structure of PPBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
8GN0
 
 | | Crystal structure of DCBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
8GN1
 
 | | Crystal structure of DBBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
9KQP
 
 | | PSI-LHCI supercomplex binding with 10 Lhcas from C. subellipsoidea | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3~{E},5~{E},7~{E})-6-methyl-8-[(6~{R})-2,2,6-trimethylcyclohexyl]octa-3,5,7-trien-2-one, ... | | Authors: | Tsai, P.-C, Kato, K, Shen, J.-R, Akita, F. | | Deposit date: | 2024-11-26 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | Structural study of the chlorophyll between Lhca8 and PsaJ in an Antarctica green algal photosystem I-LHCI supercomplex revealed by its atomic structure.
Biochim Biophys Acta Bioenerg, 1866, 2025
|
|
9KQQ
 
 | | PSI-LHCI supercomplex binding with 8 Lhcas from C. subellipsoidea | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Tsai, P.-C, Kato, K, Shen, J.-R, Akita, F. | | Deposit date: | 2024-11-26 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural study of the chlorophyll between Lhca8 and PsaJ in an Antarctica green algal photosystem I-LHCI supercomplex revealed by its atomic structure.
Biochim Biophys Acta Bioenerg, 1866, 2025
|
|
9KQB
 
 | | PSII-FCPII supercomplex from haptophyte Chrysotila roscoffensis | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | La Rocca, R, Kato, K, Tsai, P.-C, Nakajima, Y, Akita, F, Shen, J.-R. | | Deposit date: | 2024-11-25 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Structure of a photosystem II-FCPII supercomplex from a haptophyte reveals a distinct antenna organization.
Nat Commun, 16, 2025
|
|
7DZH
 
 | | intermediate of FABP with a delay time of 100 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZE
 
 | | Fabp ground state captured by XFELs | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZF
 
 | | Intermediate of FABP with a delay time of 10 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
