3SZS
 
 | |
3UFE
 
 | | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grosse, C, Himmel, S, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2011-11-01 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution
To be Published
|
|
2J80
 
 | | Structure of Citrate-bound Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | CITRATE ANION, GLYCEROL, SENSOR KINASE CITA, ... | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2006-10-18 | | Release date: | 2007-10-23 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
2J8T
 
 | | Human aldose reductase in complex with NADP and citrate at 0.82 angstrom | | Descriptor: | ALDO-KETO REDUCTASE FAMILY 1, MEMBER B1, CITRATE ANION, ... | | Authors: | Biadene, M, Hazemann, I, Cousido, A, Ginell, S, Sheldrick, G.M, Podjarny, A, Schneider, T.R. | | Deposit date: | 2006-10-27 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.82 Å) | | Cite: | The Atomic Resolution Structure of Human Aldose Reductase Reveals that Rearrangement of a Bound Ligand Allows the Opening of the Safety-Belt Loop.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3C8P
 
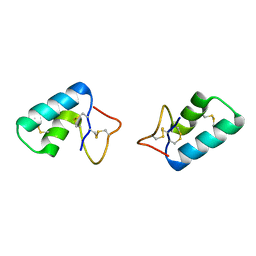 | | X-ray structure of Viscotoxin A1 from Viscum album L. | | Descriptor: | Viscotoxin A1 | | Authors: | Pal, A, Debreczeni, J.E, Sevvana, M, Gruene, T, Kahle, B, Zeeck, A, Sheldrick, G.M. | | Deposit date: | 2008-02-13 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of viscotoxins A1 and B2 from European mistletoe solved using native data alone
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3C2J
 
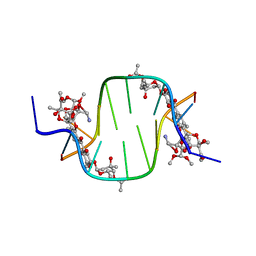 | |
3C1P
 
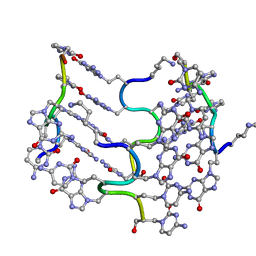 | | Crystal Structure of an alternating D-Alanyl, L-Homoalanyl PNA | | Descriptor: | Peptide Nucleic Acid DLY-HGL-AGD-LHC-AGD-LHC-CUD-LYS | | Authors: | Cuesta-Seijo, J.A, Sheldrick, G.M, Zhang, J, Diederichsen, U. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Continuous beta-turn fold of an alternating alanyl/homoalanyl peptide nucleic acid.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3DU1
 
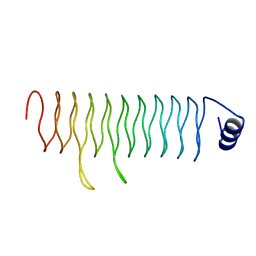 | |
3EE6
 
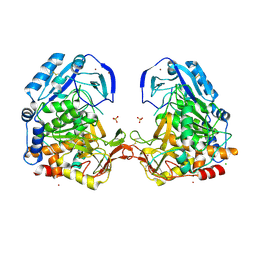 | | Crystal Structure Analysis of Tripeptidyl peptidase -I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pal, A, Kraetzner, R, Grapp, M, Gruene, T, Schreiber, K, Granborg, M, Urlaub, H, Asif, A.R, Becker, S, Gartner, J, Sheldrick, G.M, Steinfeld, R. | | Deposit date: | 2008-09-04 | | Release date: | 2008-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis
J.Biol.Chem., 284, 2009
|
|
3GO3
 
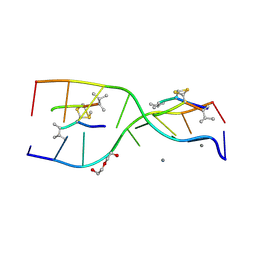 | | Interactions of an echinomycin-DNA complex with manganese(II) ions | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*AP*CP*GP*TP*AP*CP*GP*T)-3', DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pfoh, R, Cuesta-Seijo, J.A, Sheldrick, G.M. | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Interaction of an Echinomycin-DNA Complex with Manganese Ion
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3GWH
 
 | | Crystallographic Ab Initio protein solution far below atomic resolution | | Descriptor: | PHOSPHATE ION, Transcriptional antiterminator (BglG family) | | Authors: | Rodriguez, D.D, Grosse, C, Himmel, S, Gonzalez, C, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic ab initio protein structure solution below atomic resolution
Nat.Methods, 6, 2009
|
|
3JU4
 
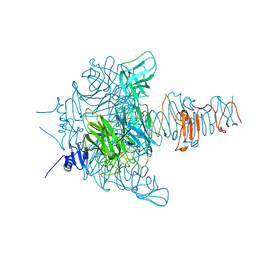 | | Crystal Structure Analysis of EndosialidaseNF at 0.98 A Resolution | | Descriptor: | CHLORIDE ION, Endo-N-acetylneuraminidase, N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Neuman, P, Gerardy-Schahn, R, Sheldrick, G.M, Ficner, R. | | Deposit date: | 2009-09-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure analysis of endosialidase NF at 0.98 A resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MBU
 
 | | Structure of a bipyridine-modified PNA duplex | | Descriptor: | 1,2-ETHANEDIOL, Bipyridine-PNA, CARBONATE ION, ... | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Du, S, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
3MBC
 
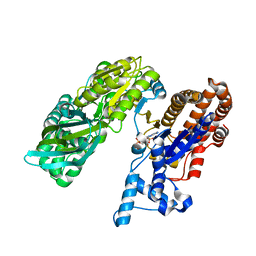 | | Crystal structure of monomeric isocitrate dehydrogenase from Corynebacterium glutamicum in complex with NADP | | Descriptor: | Isocitrate dehydrogenase [NADP], MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sidhu, N.S, Aich, S, Sheldrick, G.M, Delbaere, L.T.J. | | Deposit date: | 2010-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly NADP+-specific isocitrate dehydrogenase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3MBS
 
 | | Crystal structure of 8mer PNA | | Descriptor: | 1,2-ETHANEDIOL, Peptide Nucleic Acid | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
1A7Z
 
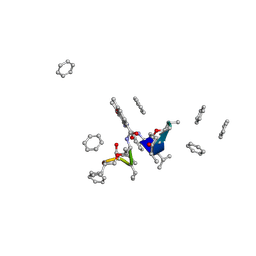 | | CRYSTAL STRUCTURE OF ACTINOMYCIN Z3 | | Descriptor: | ACTINOMYCIN Z3, BENZENE | | Authors: | Schafer, M. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
1C58
 
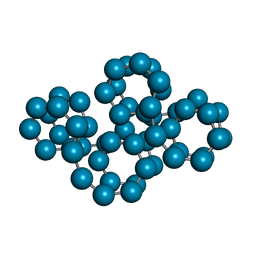 | | CRYSTAL STRUCTURE OF CYCLOAMYLOSE 26 | | Descriptor: | Cyclohexacosakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Gessler, K, Saenger, W, Nimz, O. | | Deposit date: | 1999-11-04 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | V-Amylose at atomic resolution: X-ray structure of a cycloamylose with 26 glucose residues (cyclomaltohexaicosaose).
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1E6F
 
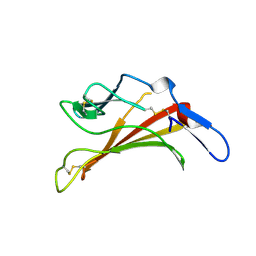 | | Human MIR-receptor, repeat 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Von Buelow, R, Rajashankar, K.R, Dauter, M, Dauter, Z, Grimme, S, Schmidt, B, Von Figura, K, Uson, I. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Locating the Anomalous Scatterer Substructures in Halide and Sulfur Phasing
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OIJ
 
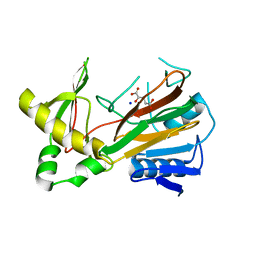 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with alphaketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, PUTATIVE ALKYLSULFATASE ATSK, SODIUM ION | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1OIH
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase | | Descriptor: | PUTATIVE ALKYLSULFATASE ATSK, SODIUM ION | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) -Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1OIK
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with fe, alphaketoglutarate and 2-ethyl-1-hexanesulfuric acid | | Descriptor: | (2R)-2-ETHYL-1-HEXANESULFONIC ACID, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1OII
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with iron and alphaketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, PUTATIVE ALKYLSULFATASE ATSK | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1PFE
 
 | | Echinomycin-(gcgtacgc)2 complex | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3', CHLORIDE ION, ... | | Authors: | Cuesta-Seijo, J.A. | | Deposit date: | 2003-05-26 | | Release date: | 2004-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of Complexes between Echinomycin and Duplex DNA.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1GQB
 
 | | HUMAN MIR-RECEPTOR, REPEAT 11 | | Descriptor: | BROMIDE ION, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Von Buelow, R, Dauter, M, Dauter, Z, Rajashankar, K.R, Grimme, S, Schmidt, B, Von Figura, K, Uson, I. | | Deposit date: | 2001-11-22 | | Release date: | 2002-12-05 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Locating the Anomalous Scatterer Substructures in Halide and Sulfur Phasing
Acta Crystallogr.,Sect.D, 59, 2003
|
|
