1LF1
 
 | | Crystal Structure of Cel5 from Alkalophilic Bacillus sp. | | Descriptor: | Cel5 | | Authors: | Shaw, A, Bott, R, Vonrhein, C, Bricogne, G, Power, S, Day, A.G. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel combination of two classic catalytic schemes.
J.Mol.Biol., 320, 2002
|
|
1LYN
 
 | |
1LIS
 
 | | THE CRYSTAL STRUCTURE OF A FERTILIZATION PROTEIN | | Descriptor: | LYSIN | | Authors: | Shaw, A, Mcree, D.E, Vacquier, V.D, Stout, C.D. | | Deposit date: | 1993-06-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of lysin, a fertilization protein.
Science, 262, 1993
|
|
2EA3
 
 | |
2CKR
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THERMOBIFIDA FUSCA ENDOGLUCANASE CEL5A (E5) E355Q IN COMPLEX WITH CELLOTETRAOSE | | Descriptor: | BENZAMIDINE, ENDOGLUCANASE E-5, SODIUM ION, ... | | Authors: | Berglund, G.I, Gualfetti, P.J, Requadt, C, Gross, L.S, Bergfors, T, Shaw, A, Saldajeno, M, Mitchinson, C, Sandgren, M. | | Deposit date: | 2006-04-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal Structure of the Catalytic Domain of Thermobifida Fusca Endoglucanase Cel5A in Complex with Cellotetraose
To be Published
|
|
2CKS
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THERMOBIFIDA FUSCA ENDOGLUCANASE CEL5A (E5) | | Descriptor: | BENZAMIDINE, ENDOGLUCANASE E-5, SODIUM ION, ... | | Authors: | Berglund, G.I, Gualfetti, P.J, Requadt, C, Gross, L.S, Bergfors, T, Shaw, A, Saldajeno, M, Mitchinson, C, Sandgren, M. | | Deposit date: | 2006-04-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Catalytic Domain of Thermobifida Fusca Endoglucanase Cel5A in Complex with Cellotetraose
To be Published
|
|
5CCP
 
 | | HISTIDINE 52 IS A CRITICAL RESIDUE FOR RAPID FORMATION OF CYTOCHROME C PEROXIDASE COMPOUND I | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brown, K, Shaw, A, Miller, M.A, Kraut, J. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I.
Biochemistry, 32, 1993
|
|
1OLQ
 
 | | The Trichoderma reesei cel12a P201C mutant, structure at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
1OLR
 
 | | The Humicola grisea Cel12A Enzyme Structure at 1.2 A Resolution | | Descriptor: | ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
1OA2
 
 | | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Day, A.G, Jones, T.A, Mitchinson, C. | | Deposit date: | 2002-12-28 | | Release date: | 2003-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability
Protein Sci., 12, 2003
|
|
1OA3
 
 | | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1-4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Day, A.G, Jones, T.A, Mitchinson, C. | | Deposit date: | 2002-12-28 | | Release date: | 2003-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability
Protein Sci., 12, 2003
|
|
1OA4
 
 | | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability | | Descriptor: | ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Day, A.G, Jones, T.A, Mitchinson, C. | | Deposit date: | 2002-12-28 | | Release date: | 2003-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability
Protein Sci., 12, 2003
|
|
1S3S
 
 | | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin containing protein) (VCP) [Contains: Valosin], p47 protein | | Authors: | Dreveny, I, Kondo, H, Uchiyama, K, Shaw, A, Zhang, X, Freemont, P.S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47.
Embo J., 23, 2004
|
|
1UU6
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH A SOAKED CELLOPENTAOSE | | Descriptor: | ENDO-BETA-1,4-GLUCANASE, TETRAETHYLENE GLYCOL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2003-12-15 | | Release date: | 2004-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1A8P
 
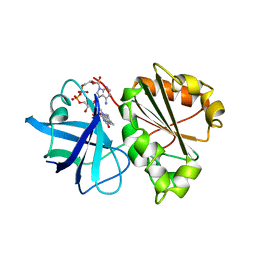 | | FERREDOXIN REDUCTASE FROM AZOTOBACTER VINELANDII | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH:FERREDOXIN OXIDOREDUCTASE | | Authors: | Prasad, G.S, Kresge, N, Muhlberg, A.B, Shaw, A, Jung, Y.S, Burgess, B.K, Stout, C.D. | | Deposit date: | 1998-03-28 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of NADPH:ferredoxin reductase from Azotobacter vinelandii.
Protein Sci., 7, 1998
|
|
1W2U
 
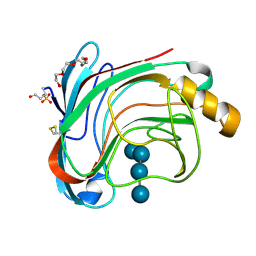 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH A SOAKED THIO CELLOTETRAOSE | | Descriptor: | ENDOGLUCANASE, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1UU4
 
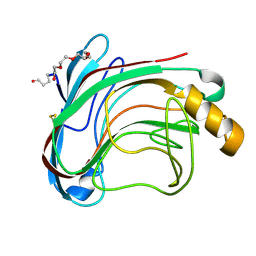 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH CELLOBIOSE | | Descriptor: | ENDO-BETA-1,4-GLUCANASE, TETRAETHYLENE GLYCOL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2003-12-15 | | Release date: | 2004-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1DCC
 
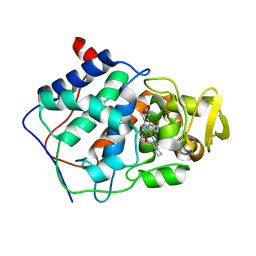 | |
1E32
 
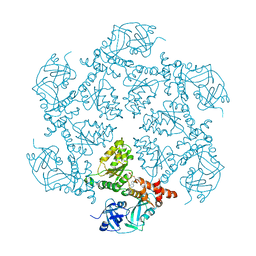 | | Structure of the N-Terminal domain and the D1 AAA domain of membrane fusion ATPase p97 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, P97 | | Authors: | Zhang, X, Shaw, A, Bates, P.A, Gorman, M.A, Kondo, H, Dokurno, P, Leonard M, G, Sternberg, J.E, Freemont, P.S. | | Deposit date: | 2000-06-05 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Aaa ATPase P97
Mol.Cell, 6, 2000
|
|
1UU5
 
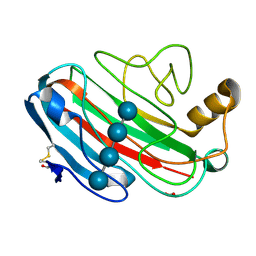 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A SOAKED WITH CELLOTETRAOSE | | Descriptor: | ACETATE ION, ENDO-BETA-1,4-GLUCANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2003-12-15 | | Release date: | 2004-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1I42
 
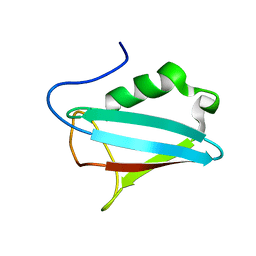 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 | | Descriptor: | P47 | | Authors: | Yuan, X, Shaw, A, Zhang, X, Kondo, H, Lally, J, Freemont, P.S, Matthews, S. | | Deposit date: | 2001-02-19 | | Release date: | 2001-08-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
1H8V
 
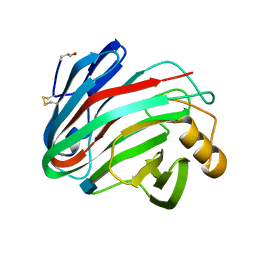 | | The X-ray Crystal Structure of the Trichoderma reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Shaw, A, Ropp, T.H, Wu, S, Bott, R, Cameron, A.D, Stahlberg, J, Mitchinson, C, Jones, T.A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-Ray Crystal Structure of the Trichoderma Reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution
J.Mol.Biol., 308, 2001
|
|
1JRU
 
 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 (ENERGY MINIMISED AVERAGE) | | Descriptor: | p47 protein | | Authors: | Yuan, X.M, Shaw, A, Zhang, X.D, Kondo, H, Lally, J, Freemont, P.S, Matthews, S.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
5WJN
 
 | | Crystal Structure of HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, GTS3 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WJL
 
 | | Crystal Structure of HLA-A*11:01 with GTS1 peptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GTS1 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
