3BN6
 
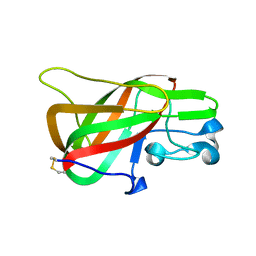 | | Crystal Structure of the C2 Domain of Bovine Lactadherin at 1.67 Angstrom Resolution | | Descriptor: | Lactadherin | | Authors: | Shao, C, Novakovic, V.A, Head, J.F, Seaton, B.A, Gilbert, G.E. | | Deposit date: | 2007-12-13 | | Release date: | 2007-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of lactadherin C2 domain at 1.7A resolution with mutational and computational analyses of its membrane-binding motif.
J.Biol.Chem., 283, 2008
|
|
2HYU
 
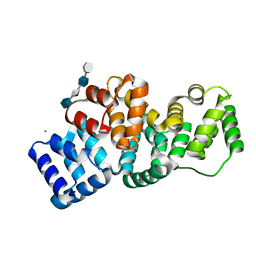 | | Human Annexin A2 with heparin tetrasaccharide bound | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Annexin A2, CALCIUM ION | | Authors: | Shao, C, Head, J.F, Seaton, B.A. | | Deposit date: | 2006-08-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic Analysis of Calcium-dependent Heparin Binding to Annexin A2.
J.Biol.Chem., 281, 2006
|
|
2HYW
 
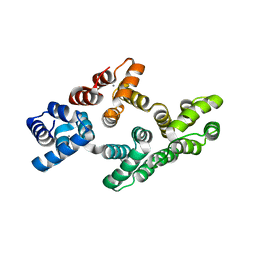 | | Human Annexin A2 with Calcium bound | | Descriptor: | Annexin A2, CALCIUM ION | | Authors: | Shao, C, Head, J.F, Seaton, B.A. | | Deposit date: | 2006-08-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Analysis of Calcium-dependent Heparin Binding to Annexin A2.
J.Biol.Chem., 281, 2006
|
|
2HYV
 
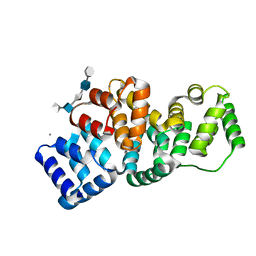 | | Human Annexin A2 with heparin hexasaccharide bound | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Annexin A2, CALCIUM ION | | Authors: | Shao, C, Head, J.F, Seaton, B.A. | | Deposit date: | 2006-08-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic analysis of calcium-dependent heparin binding to annexin A2.
J.Biol.Chem., 281, 2006
|
|
2OR2
 
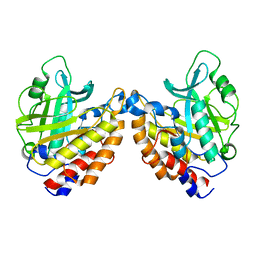 | | Structure of the W47A/W242A Mutant of Bacterial Phosphatidylinositol-Specific Phospholipase C | | Descriptor: | 1-phosphatidylinositol phosphodiesterase | | Authors: | Shao, C, Shi, X, Wehbi, H, Zambonelli, C, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2007-02-01 | | Release date: | 2007-02-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dimer structure of an interfacially impaired phosphatidylinositol-specific phospholipase C.
J.Biol.Chem., 282, 2007
|
|
4OKY
 
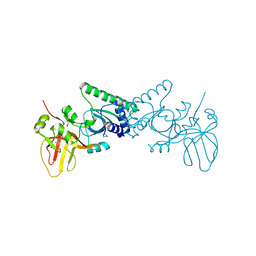 | | Crystal structure of PvuRts1I, a 5-hydroxymethylcytosine DNA restriction endonuclease | | Descriptor: | Restriction endonuclease PvuRts1 I | | Authors: | Wang, C.L, Shao, C, Zang, J.Y. | | Deposit date: | 2014-01-23 | | Release date: | 2014-09-10 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the substrate selectivity of PvuRts1I, a 5-hydroxymethylcytosine DNA restriction endonuclease
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5J6P
 
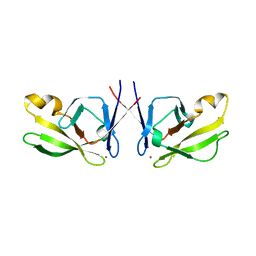 | | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe | | Descriptor: | Kinetochore protein mis18, ZINC ION | | Authors: | Wang, C, Shao, C, Zhang, M, Zhang, X, Zang, J. | | Deposit date: | 2016-04-05 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe
To Be Published
|
|
3EA3
 
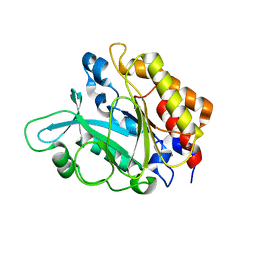 | | Crystal Structure of the Y246S/Y247S/Y248S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, MANGANESE (II) ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Modulation of Bacillus thuringiensis Phosphatidylinositol-specific Phospholipase C Activity by Mutations in the Putative Dimerization Interface.
J.Biol.Chem., 284, 2009
|
|
3EA2
 
 | | Crystal Structure of the Myo-inositol bound Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
3EA1
 
 | | Crystal Structure of the Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
5CB3
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5CMS
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, SULFATE ION | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
4IFS
 
 | | Crystal structure of the hSSRP1 Middle domain | | Descriptor: | CHLORIDE ION, FACT complex subunit SSRP1 | | Authors: | Zhang, W.J, Zeng, F.X, Shao, C, Liu, Y.W, Niu, L.W, Li, X, Teng, M.K. | | Deposit date: | 2012-12-15 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the hSSRP1 Middle domain
To be Published
|
|
5QD7
 
 | | Crystal structure of BACE complex with BMC014 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({1-[3-(1-methylethyl)phenyl]cyclopropyl}amino)ethyl]-19-(methoxymethyl)-11-oxa-3,16-diazatric yclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5Q1B
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{[(2S)-2-cyclohexyl-2-{5,6-difluoro-2-[4-(1,3-thiazol-2-yl)phenyl]-1H-benzimidazol-1-yl}acetyl]amino}benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-[(3,4-dimethoxyphenyl)-(4-methylphenyl)sulfonyl-amino]-N-(2,4-dimethylpentan-3-yl)ethanamide, Bile acid receptor, COACTIVATOR PEPTIDE PGC-1A PPAR GAMMA COACTIVATOR | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Q
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, ethyl 4-({2-phenyl-5-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-2H-pyrazolo[4,3-c]pyridine-3-carbonyl}amino)benzoate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q13
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-[6-chloro-2-(4-chlorophenyl)-5-fluoro-1H-benzimidazol-1-yl]-N-cyclohexyl-2-[(2S)-oxan-2-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0M
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-{[(3beta,5beta,14beta,17alpha)-3-hydroxy-24-oxocholan-24-yl]amino}benzene-1,3-dicarboxylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0W
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-({5-bromo-1'-[(2-chlorophenyl)sulfonyl]-2-oxospiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, cDNA FLJ76652, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q12
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-(2,6-difluorophenyl)-N-(2,6-dimethylphenyl)-5-methylimidazo[1,2-a]pyridin-3-amine, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1G
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2E)-N-cyclohexyl-N-(cyclohexylcarbamoyl)-3-(4-fluorophenyl)prop-2-enamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0O
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-{2-[4-(benzenecarbonyl)phenyl]-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, CHLORIDE ION, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q14
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}-3-fluorobenzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
