3J8X
 
 | | High-resolution structure of no-nucleotide kinesin on microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
3J8Y
 
 | | High-resolution structure of ATP analog-bound kinesin on microtubules | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
5YGH
 
 | | Crystal Structure of the Capsid Protein from Zika Virus | | Descriptor: | Capsid protein | | Authors: | Shang, Z, Song, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-09-23 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Crystal Structure of the Capsid Protein from Zika Virus.
J. Mol. Biol., 430, 2018
|
|
6PWB
 
 | | Rigid body fitting of flagellin FlaB, and flagellar coiling proteins, FcpA and FcpB, into a 10 Angstrom structure of the asymmetric flagellar filament purified from Leptospira biflexa Patoc WT cells resolved via subtomogram averaging | | Descriptor: | Flagellar coiling protein A (FcpA), Flagellar coiling protein B (FcpB), Flagellin B1 (FlaB1) | | Authors: | Gibson, K.H, Sindelar, C.V, Trajtenberg, F, Buschiazzo, A, San Martin, F, Mechaly, A. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.83 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the leptospira spirochete.
Elife, 9, 2020
|
|
7VWY
 
 | |
7VWZ
 
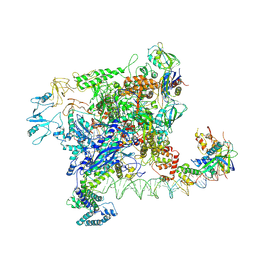 | | Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-12 | | Release date: | 2022-06-08 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription activation by Rob, a pleiotropic AraC/XylS family regulator.
Nucleic Acids Res., 50, 2022
|
|
7EQL
 
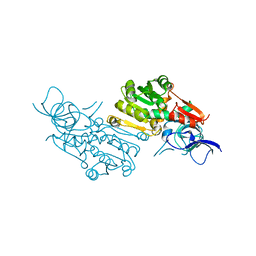 | | Crystal structure of (+)-pulegone reductase from Mentha piperita | | Descriptor: | (+)-pulegone reductase | | Authors: | Lin, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Functional Characterization and Structural Insights Into Stereoselectivity of Pulegone Reductase in Menthol Biosynthesis.
Front Plant Sci, 12, 2021
|
|
7C7E
 
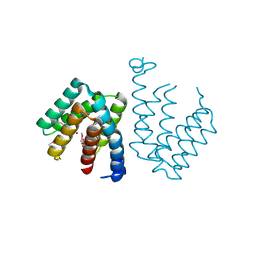 | | Crystal structure of C terminal domain of Escherichia coli DgoR | | Descriptor: | Putative DNA-binding transcriptional regulator, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Lin, W. | | Deposit date: | 2020-05-25 | | Release date: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural and Functional Analyses of the Transcription Repressor DgoR From Escherichia coli Reveal a Divalent Metal-Containing D-Galactonate Binding Pocket.
Front Microbiol, 11, 2020
|
|
6ILM
 
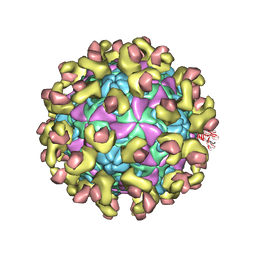 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 7.4 | | Descriptor: | Beta-2-microglobulin, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILO
 
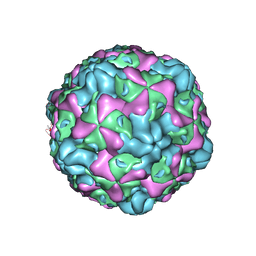 | | Cryo-EM structure of empty Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILL
 
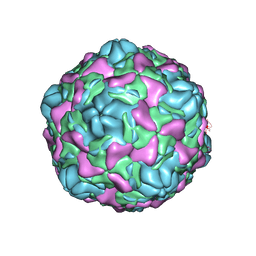 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILJ
 
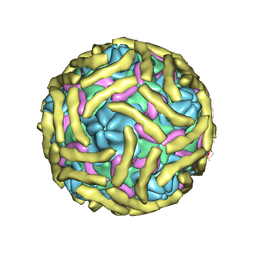 | | Cryo-EM structure of Echovirus 6 complexed with its attachment receptor CD55 at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-18 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILK
 
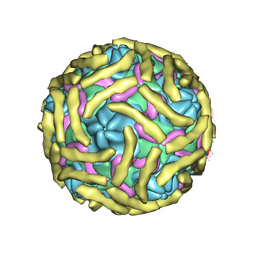 | | Cryo-EM structure of Echovirus 6 complexed with its attachment receptor CD55 at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-18 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILP
 
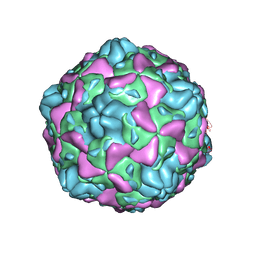 | | Cryo-EM structure of full Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILN
 
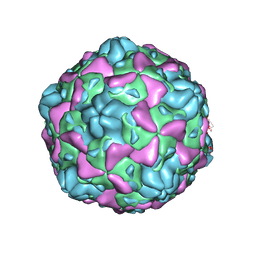 | | Cryo-EM structure of full Echovirus 6 particle at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
7DY6
 
 | |
7YTV
 
 | |
6LOI
 
 | |
7WMX
 
 | |
7WMZ
 
 | |
7WMY
 
 | |
7WMW
 
 | |
