5TJW
 
 | | Influenza A virus Nucleoprotein in Complex with Inhibitory Nanobody | | Descriptor: | NP-specific inhibitory VHH, Nucleoprotein | | Authors: | Hanke, L, Knockenhauer, K.E, Ploegh, H.L, Schwartz, T.U. | | Deposit date: | 2016-10-05 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.231 Å) | | Cite: | The Antiviral Mechanism of an Influenza A Virus Nucleoprotein-Specific Single-Domain Antibody Fragment.
MBio, 7, 2016
|
|
5UKB
 
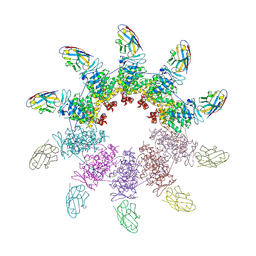 | | VSV N PROTEIN IN COMPLEX WITH INHIBITORY NANOBODY 1004 | | Descriptor: | Anti-vesicular stomatitis virus N VHH, Nucleocapsid, RNA (45-MER) | | Authors: | Hanke, L, Knockenhauer, K.E, Ploegh, H.L, Schwartz, T.U. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (5.473 Å) | | Cite: | Vesicular stomatitis virus N protein-specific single-domain antibody fragments inhibit replication.
EMBO Rep., 18, 2017
|
|
5UK4
 
 | | VESICULAR STOMATITS VIRUS N PROTEIN IN COMPLEX WITH INHIBITORY NANOBODY 1307 | | Descriptor: | Anti-vesicular stomatitis virus N VHH, Nucleoprotein, RNA (45-MER) | | Authors: | Hanke, L, Knockenhauer, K.E, Ploegh, H.L, Schwartz, T.U. | | Deposit date: | 2017-01-19 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Vesicular stomatitis virus N protein-specific single-domain antibody fragments inhibit replication.
EMBO Rep., 18, 2017
|
|
4FCC
 
 | | Glutamate dehydrogenase from E. coli | | Descriptor: | Glutamate dehydrogenase | | Authors: | Bilokapic, S, Schwartz, T.U. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FHM
 
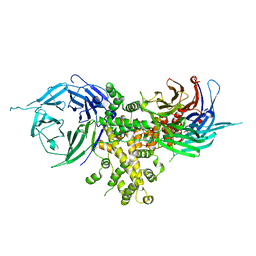 | |
4FHL
 
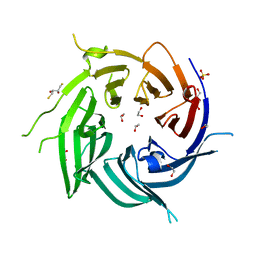 | | Nucleoporin Nup37 from Schizosaccharomyces pombe | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Bilokapic, S, Schwartz, T.U. | | Deposit date: | 2012-06-06 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FHN
 
 | |
4I0O
 
 | |
5I2C
 
 | |
5J1S
 
 | | TorsinA-LULL1 complex, H. sapiens, bound to VHH-BS2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Demircioglu, F.E, Schwartz, T.U. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia.
Elife, 5, 2016
|
|
5JJ7
 
 | | Fic-1 (aa134 - 508 E274G) from C. elegans | | Descriptor: | Adenosine monophosphate-protein transferase FICD homolog, SULFATE ION | | Authors: | Cruz, V.E, Schwartz, T.U. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.749 Å) | | Cite: | Fic-1 (aa134 - 508 E274G) from C. elegans
To be published
|
|
5J1T
 
 | | TorsinAdeltaE-LULL1 complex, H. sapiens, bound to VHH-BS2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Demircioglu, F.E, Schwartz, T.U. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia.
Elife, 5, 2016
|
|
5JJ6
 
 | |
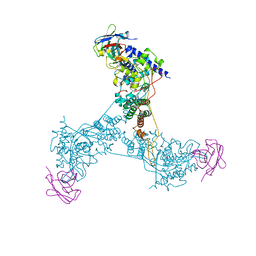
6X08
 
 | | Nup85-Seh1 from S. cerevisiae bound by VHH-SAN2 | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1, VHH-SAN2 | | Authors: | Nordeen, S.A, Schwartz, T.U. | | Deposit date: | 2020-05-15 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | A nanobody suite for yeast scaffold nucleoporins provides details of the nuclear pore complex structure.
Nat Commun, 11, 2020
|
|
6WMF
 
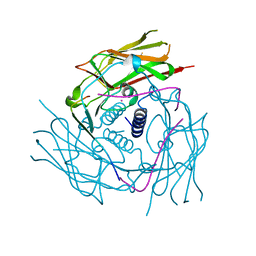 | | Human Sun2-KASH5 complex | | Descriptor: | POTASSIUM ION, Protein KASH5, SUN domain-containing protein 2 | | Authors: | Cruz, V.E, Schwartz, T.U. | | Deposit date: | 2020-04-21 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of Different LINC Complexes Reveals Distinct Binding Modes.
J.Mol.Biol., 432, 2020
|
|
6WME
 
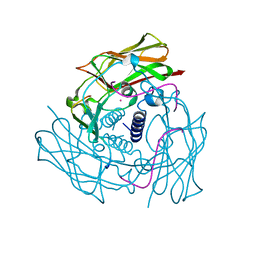 | | Human Sun2-KASH3 complex | | Descriptor: | Nesprin-3, POTASSIUM ION, SUN domain-containing protein 2 | | Authors: | Cruz, V.E, Schwartz, T.U. | | Deposit date: | 2020-04-21 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Analysis of Different LINC Complexes Reveals Distinct Binding Modes.
J.Mol.Biol., 432, 2020
|
|
6WMG
 
 | | Human Sun2 (500-717) | | Descriptor: | POTASSIUM ION, SUN domain-containing protein 2 | | Authors: | Cruz, V.E, Schwartz, T.U. | | Deposit date: | 2020-04-21 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Different LINC Complexes Reveals Distinct Binding Modes.
J.Mol.Biol., 432, 2020
|
|
6X05
 
 | |
6WMD
 
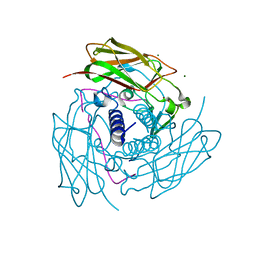 | | Human Sun2-KASH4 complex | | Descriptor: | MAGNESIUM ION, Nesprin-4, SUN domain-containing protein 2 | | Authors: | Cruz, V.E, Schwartz, T.U. | | Deposit date: | 2020-04-21 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Different LINC Complexes Reveals Distinct Binding Modes.
J.Mol.Biol., 432, 2020
|
|
6X07
 
 | |
6X04
 
 | |
6X06
 
 | |
6X02
 
 | |
6X03
 
 | |
3GJ0
 
 | | Crystal structure of human RanGDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Partridge, J.R, Schwartz, T.U. | | Deposit date: | 2009-03-07 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic and Biochemical Analysis of the Ran-binding Zinc Finger Domain.
J.Mol.Biol., 391, 2009
|
|
