1UWL
 
 | |
1W1U
 
 | |
1W4R
 
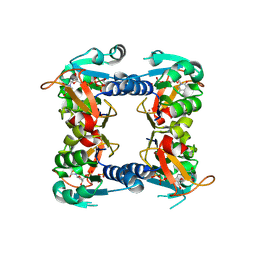 | | Structure of a type II thymidine kinase with bound dTTP | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Birringer, M.S, Claus, M.T, Folkers, G, Kloer, D.P, Schulz, G.E, Scapozza, L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-02-01 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of a Type II Thymidine Kinase with Bound Dttp
FEBS Lett., 579, 2005
|
|
1W27
 
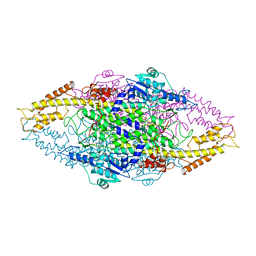 | | Phenylalanine ammonia-lyase (PAL) from Petroselinum crispum | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PHENYLALANINE AMMONIA-LYASE 1 | | Authors: | Ritter, H, Schulz, G.E. | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Entrance Into the Phenylpropanoid Metabolism Catalyzed by Phenylalanine Ammonia-Lyase
Plant Cell, 16, 2004
|
|
1X7O
 
 | |
1X7P
 
 | |
1DZX
 
 | |
1DZW
 
 | |
1E1N
 
 | |
1DZV
 
 | |
1DZU
 
 | |
1E1L
 
 | |
1DZY
 
 | |
1DZZ
 
 | |
1E1K
 
 | |
1E1M
 
 | |
1E4Y
 
 | |
1EB4
 
 | |
1E4V
 
 | |
1GKM
 
 | |
1GK2
 
 | |
1GK3
 
 | |
1GKY
 
 | |
1GES
 
 | |
1GEU
 
 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
