3GVN
 
 | | The 1.2 Angstroem crystal structure of an E.coli tRNASer acceptor stem microhelix reveals two magnesium binding sites | | Descriptor: | 5'-R(*CP*CP*UP*CP*AP*CP*C)-3', 5'-R(*GP*GP*UP*GP*AP*GP*G)-3', MAGNESIUM ION | | Authors: | Eichert, A, Furste, J.P, Schreiber, A, Perbandt, M, Betzel, C, Erdmann, V.A, Forster, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2A crystal structure of an E. coli tRNASer)acceptor stem microhelix reveals two magnesium binding sites.
Biochem.Biophys.Res.Commun., 386, 2009
|
|
2VUQ
 
 | | Crystal structure of a human tRNAGly acceptor stem microhelix (derived from the gene sequence DG9990) at 1.18 Angstroem resolution | | Descriptor: | 5'-R(*CP*CP*AP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*UP*GP*GP)-3' | | Authors: | Eichert, A, Perbandt, M, Schreiber, A, Fuerste, J.P, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2008-05-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal Structure of the Human Trnagly Microhelix Isoacceptor G9990 at 1.18 A Resolution
Biochem.Biophys.Res.Commun., 380, 2009
|
|
2W89
 
 | | Crystal structure of the E.coli tRNAArg aminoacyl stem issoacceptor RR-1660 at 2.0 Angstroem resolution | | Descriptor: | 5'-R(*CP*GP*GP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*CP*CP*GP)-3', GLYCEROL | | Authors: | Eichert, A, Schreiber, A, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E. Coli tRNA(Arg) Aminoacyl Stem Isoacceptor Rr-1660 at 2.0 A Resolution.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
7P5Z
 
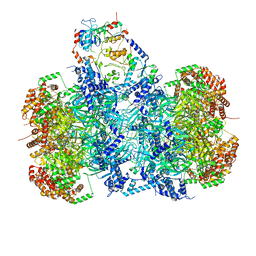 | | Structure of a DNA-loaded MCM double hexamer engaged with the Dbf4-dependent kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 7, ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P30
 
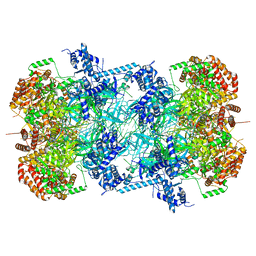 | | 3.0 A resolution structure of a DNA-loaded MCM double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (53-MER), ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
