1A8G
 
 | | HIV-1 PROTEASE IN COMPLEX WITH SDZ283-910 | | Descriptor: | HIV-1 PROTEASE, benzyl [(1R)-1-({(1S,2S,3S)-1-benzyl-2-hydroxy-4-({(1S)-1-[(2-hydroxy-4-methoxybenzyl)carbamoyl]-2-methylpropyl}amino)-3-[(4-methoxybenzyl)amino]-4-oxobutyl}carbamoyl)-2,2-dimethylpropyl]carbamate | | Authors: | Kallen, J, Billich, A, Scholz, D, Auer, M, Kungl, A. | | Deposit date: | 1998-03-24 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure and conformational dynamics of the HIV-1 protease in complex with the inhibitor SDZ283-910: agreement of time-resolved spectroscopy and molecular dynamics simulations.
J.Mol.Biol., 286, 1999
|
|
8P3E
 
 | | Crystal structure of glucocerebrosidase in complex with allosteric activator | | Descriptor: | 2-[[3-[(4-chlorophenyl)carbamoyl]phenyl]sulfonylamino]benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-05-17 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of ss-Glucocerebrosidase Activators for Glucosylceramide hydrolysis.
Chemmedchem, 19, 2024
|
|
8P41
 
 | | Crystal structure of glucocerebrosidase in complex with allosteric activator | | Descriptor: | 2-[2-[[3,5-bis(trifluoromethyl)phenyl]methylsulfanyl]ethanoylamino]-5-chloranyl-benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of ss-Glucocerebrosidase Activators for Glucosylceramide hydrolysis.
Chemmedchem, 19, 2024
|
|
5OQV
 
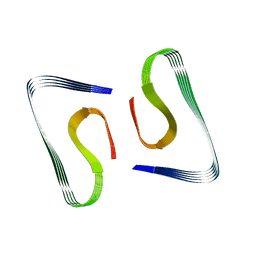 | | Near-atomic resolution fibril structure of complete amyloid-beta(1-42) by cryo-EM | | Descriptor: | Amyloid beta A4 protein | | Authors: | Gremer, L, Schoelzel, D, Schenk, C, Reinartz, E, Labahn, J, Ravelli, R, Tusche, M, Lopez-Iglesias, C, Hoyer, W, Heise, H, Willbold, D, Schroeder, G.F. | | Deposit date: | 2017-08-14 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Fibril structure of amyloid-beta (1-42) by cryo-electron microscopy.
Science, 358, 2017
|
|
