5CNC
 
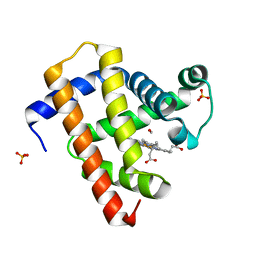 | | Ultrafast dynamics in myoglobin: 0.6 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
1LFK
 
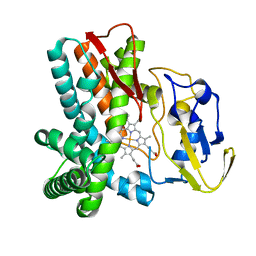 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | Descriptor: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | Deposit date: | 2002-04-11 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
3TEO
 
 | | APO Form of carbon disulfide hydrolase (selenomethionine form) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, Carbon disulfide hydrolase | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
3TEN
 
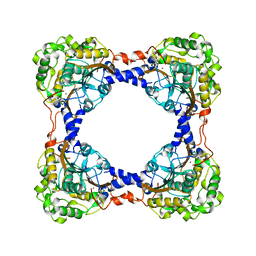 | | Holo form of carbon disulfide hydrolase | | Descriptor: | CS2 hydrolase, ZINC ION | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
1USE
 
 | | human VASP tetramerisation domain | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Kuhnel, K, Jarchau, T, Wolf, E, Schlichting, I, Walter, U, Wittinghofer, A, Strelkov, S.V. | | Deposit date: | 2003-11-21 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Vasp Tetramerization Domain is a Right-Handed Coiled Coil Based on a 15-Residue Repeat
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2J9X
 
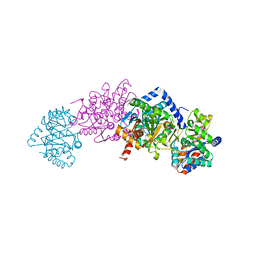 | | Tryptophan Synthase in complex with GP, alpha-D,L-glycerol-phosphate, Cs, pH6.5 - alpha aminoacrylate form - (GP)E(A-A) | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ngo, H, Kimmich, N, Harris, R, Niks, D, Blumenstein, L, Kulik, V, Barends, T.R, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-11-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Regulation of Substrate Channeling in Tryptophan Synthase: Modulation of the L-Serine Reaction in Stage I of the Beta-Reaction by Alpha-Site Ligands.
Biochemistry, 46, 2007
|
|
4QX3
 
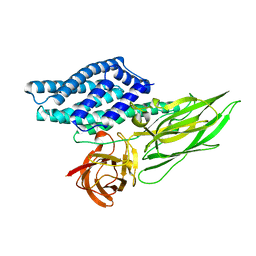 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX2
 
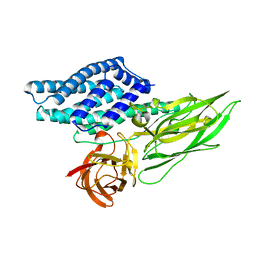 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ET9
 
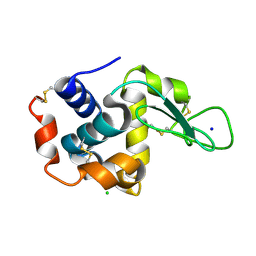 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETD
 
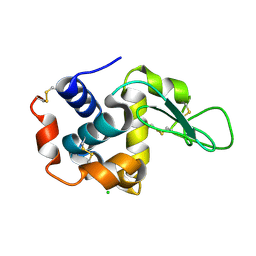 | | Lysozyme, room-temperature, rotating anode, 0.0026 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4QX1
 
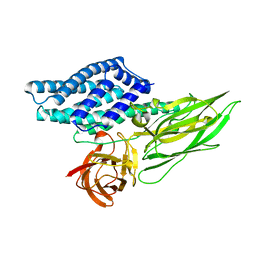 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ETA
 
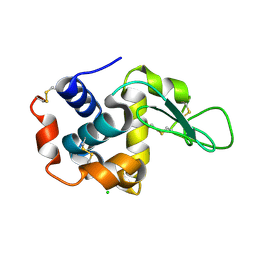 | | Lysozyme, room temperature, 400 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4XOU
 
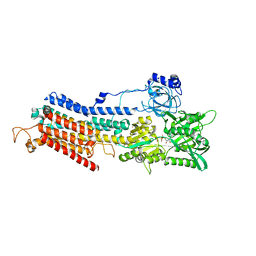 | | Crystal structure of the SR Ca2+-ATPase in the Ca2-E1-MgAMPPCP form determined by serial femtosecond crystallography using an X-ray free-electron laser. | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Nass, K, Drachmann, N.D, Markvardsen, A.J, Gutmann, M.J, Barends, T.R.M, Mattle, D, Shoeman, R.L, Doak, R.B, Boutet, S, Messerschmidt, M, Seibert, M.M, Williams, G.J, Foucar, L, Reinhard, L, Sitsel, O, Gregersen, J.L, Clausen, J.D, Boesen, T, Gotfryd, K, Wang, K.-T, Olesen, C, Moller, J.V, Nissen, P, Schlichting, I. | | Deposit date: | 2015-01-16 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of P-type ATPase-ligand complexes using an X-ray free-electron laser.
Iucrj, 2, 2015
|
|
3GFX
 
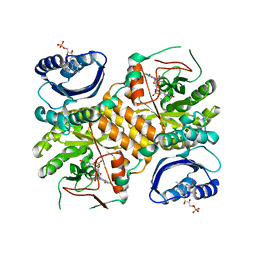 | | Klebsiella pneumoniae BlrP1 pH 4.5 calcium/cy-diGMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Barends, T, Hartmann, E, Griese, J, Beitlich, T, Kirienko, N, Ryjenkov, D, Reinstein, J, Shoeman, R, Gomelsky, M, Schlichting, I. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase.
Nature, 459, 2009
|
|
3GG1
 
 | | Klebsiella pneumoniae BlrP1 pH 8.0 calcium/cy-diGMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Barends, T, Hartmann, E, Griese, J, Beitlich, T, Kirienko, N, Ryjenkov, D, Reinstein, J, Shoeman, R, Gomelsky, M, Schlichting, I. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase.
Nature, 459, 2009
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
8BKH
 
 | |
8BKN
 
 | |
4N5R
 
 | | Hen egg-white lysozyme phased using free-electron laser data | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme C | | Authors: | Barends, T.R.M, Foucar, L, Botha, S, Doak, R.B, Shoeman, R.L, Nass, K, Koglin, J.E, Williams, G.J, Boutet, S, Messerschmidt, M, Schlichting, I. | | Deposit date: | 2013-10-10 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein crystal structure determination from X-ray free-electron laser data.
Nature, 505, 2014
|
|
4QX0
 
 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1O76
 
 | | CYANIDE COMPLEX OF P450CAM FROM PSEUDOMONAS PUTIDA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYANIDE ION, ... | | Authors: | Fedorov, R, Ghosh, D, Schlichting, I. | | Deposit date: | 2002-10-23 | | Release date: | 2002-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Cyanide Complexes of P450Cam and the Oxygenase Domain of Inducible Nitric Oxide Synthase-Structural Models of the Short-Lived Oxygen Complexes
Arch.Biochem.Biophys., 409, 2003
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | Descriptor: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS6
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | NICKEL (II) ION, PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS4
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
