1BGG
 
 | | GLUCOSIDASE A FROM BACILLUS POLYMYXA COMPLEXED WITH GLUCONATE | | Descriptor: | BETA-GLUCOSIDASE A, D-gluconic acid | | Authors: | Sanz-Aparicio, J, Hermoso, J, Martinez-Ripoll, M, Polaina, J. | | Deposit date: | 1997-05-12 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
1TR1
 
 | | CRYSTAL STRUCTURE OF E96K MUTATED BETA-GLUCOSIDASE A FROM BACILLUS POLYMYXA, AN ENZYME WITH INCREASED THERMORESISTANCE | | Descriptor: | BETA-GLUCOSIDASE A, GLYCEROL | | Authors: | Sanz-Aparicio, J, Hermoso, J.A, Martinez-Ripoll, M, Gonzalez-Perez, B, Polaina, J. | | Deposit date: | 1998-03-12 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
1BGA
 
 | |
1E4I
 
 | | 2-deoxy-2-fluoro-beta-D-glucosyl/enzyme intermediate complex of the beta-glucosidase from Bacillus polymyxa | | Descriptor: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, BETA-GLUCOSIDASE | | Authors: | Sanz-Aparicio, J, Gonzalez, B, Hermoso, J.A, Arribas, J.C, Canada, F.J, Polaina, J. | | Deposit date: | 2000-07-06 | | Release date: | 2001-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Increased Resistance to Thermal Denaturation Induced by Single Amino Acid Substitution in the Sequence of Beta-Glucosidase a from Bacillus Polymyxa.
Proteins: Struct.,Funct., Genet., 33, 1998
|
|
1AZD
 
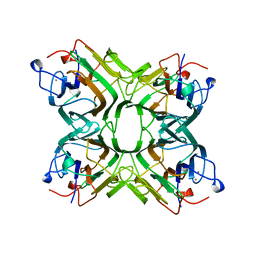 | | CONCANAVALIN FROM CANAVALIA BRASILIENSIS | | Descriptor: | CALCIUM ION, CONBR, MANGANESE (II) ION | | Authors: | Sanz-Aparicio, J, Hermoso, J, Grangeiro, T.B, Calvete, J.J, Cavada, B.S. | | Deposit date: | 1997-11-16 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of Canavalia brasiliensis lectin suggests a correlation between its quaternary conformation and its distinct biological properties from Concanavalin A.
FEBS Lett., 405, 1997
|
|
3EMZ
 
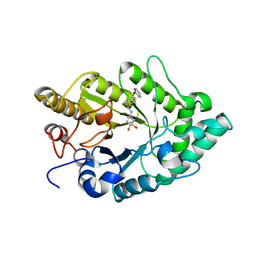 | | Crystal structure of xylanase XynB from Paenibacillus barcinonensis complexed with a conduramine derivative | | Descriptor: | (1S,2S,3R,6R)-6-[(4-phenoxybenzyl)amino]cyclohex-4-ene-1,2,3-triol, Endo-1,4-beta-xylanase | | Authors: | Sanz-Aparicio, J, Isorna, P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
3EMQ
 
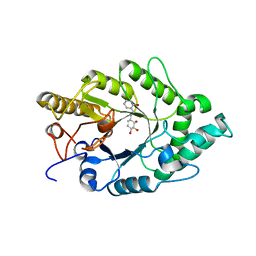 | | Crystal structure of xilanase XynB from Paenibacillus barcelonensis complexed with an inhibitor | | Descriptor: | (1S,2S,3R,6R)-6-[(2-hydroxybenzyl)amino]cyclohex-4-ene-1,2,3-triol, Endo-1,4-beta-xylanase | | Authors: | Sanz-Aparicio, J, Isorna, P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
3EMC
 
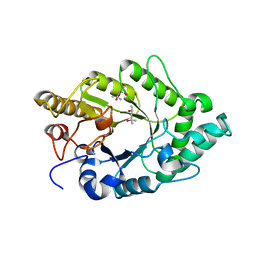 | | Crystal structure of XynB, an intracellular xylanase from Paenibacillus barcinonensis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Endo-1,4-beta-xylanase | | Authors: | Sanz-Aparicio, J, Isorna, P, Gonzalez, B. | | Deposit date: | 2008-09-24 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
3KF5
 
 | | Structure of invertase from Schwanniomyces occidentalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Invertase | | Authors: | Sanz-Aparicio, J, Polo, A. | | Deposit date: | 2009-10-27 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and kinetic analysis of Schwanniomyces occidentalis invertase reveals a new oligomerization pattern and the role of its supplementary domain in substrate binding
J.Biol.Chem., 285, 2010
|
|
3KF3
 
 | |
6FJE
 
 | |
4W8L
 
 | | Structure of GH10 from Paenibacillus barcinonensis | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase C, GLYCEROL | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2014-08-25 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring Multimodularity in Plant Cell Wall Deconstruction: STRUCTURAL AND FUNCTIONAL ANALYSIS OF Xyn10C CONTAINING THE CBM22-1-CBM22-2 TANDEM.
J.Biol.Chem., 290, 2015
|
|
4UPU
 
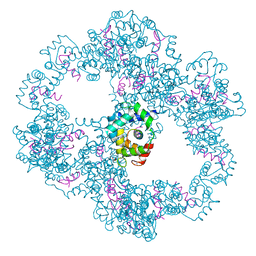 | | Crystal structure of IP3 3-K calmodulin binding region in complex with Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Franco-Echevarria, E, Banos-Sanz, J.I, Monterroso, B, Round, A, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A New Calmodulin Binding Motif for Inositol 1,4,5-Trisphosphate 3-Kinase Regulation.
Biochem.J., 463, 2014
|
|
1UMY
 
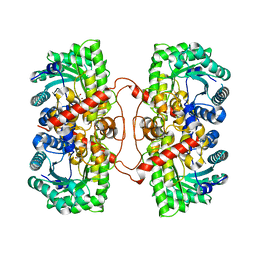 | | BHMT from rat liver | | Descriptor: | BETA-MERCAPTOETHANOL, Betaine--homocysteine S-methyltransferase 1, ZINC ION | | Authors: | Gonzalez, B, Pajares, M.A, Sanz-Aparicio, J. | | Deposit date: | 2003-09-02 | | Release date: | 2004-05-06 | | Last modified: | 2025-12-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat liver betaine homocysteine s-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding.
J. Mol. Biol., 338, 2004
|
|
8PC7
 
 | |
1E62
 
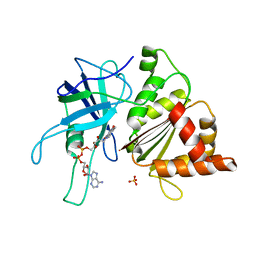 | | Ferredoxin:NADP+ reductase mutant with Lys 75 replaced by Arg (K75R) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2000-08-07 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
5K6O
 
 | |
5K6M
 
 | |
5K6L
 
 | |
7NL2
 
 | | Structure of Xyn11 from Pseudothermotoga thermarum | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-xylanase, GLYCEROL | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phylogenetic, functional and structural characterization of a GH10 xylanase active at extreme conditions of temperature and alkalinity
Comput Struct Biotechnol J, 19, 2021
|
|
1UYQ
 
 | |
4EQV
 
 | |
5ANN
 
 | | Structure of fructofuranosidase from Xanthophyllomyces dendrorhous | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-FRUCTOFURANOSIDASE, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2015-09-07 | | Release date: | 2016-02-10 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Analysis of Beta-Fructofuranosidase from Xanthophyllomyces Dendrorhous Reveals Unique Features and the Crucial Role of N-Glycosylation in Oligomerization and Activity
J.Biol.Chem., 291, 2016
|
|
1QM4
 
 | | Methionine Adenosyltransferase Complexed with a L-Methionine Analogue | | Descriptor: | L-2-AMINO-4-METHOXY-CIS-BUT-3-ENOIC ACID, MAGNESIUM ION, METHIONINE ADENOSYLTRANSFERASE, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The Crystal Structure of Tetrameric Methionine Adenosyltransferase from Rat Liver Reveals the Methionine-Binding Site
J.Mol.Biol., 300, 2000
|
|
1H85
 
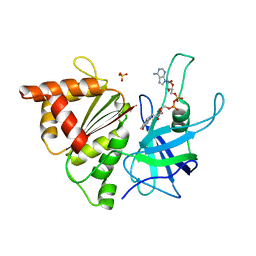 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH VAL 136 REPLACED BY LEU (V136L) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a Cluster of Hydrophobic Residues Near the Fad Cofactor in Anabaena Pcc 7119 Ferredoxin-Nadp+ Reductase for Optimal Complex Formation and Electron Transfer to Ferredoxin
J.Biol.Chem., 276, 2001
|
|
