5C6X
 
 | | Crystal structure of C-As lyase with Co(II) | | Descriptor: | COBALT (II) ION, GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of C-As lyase with Co(II)
To Be Published
|
|
5CB9
 
 | | Crystal structure of C-As lyase with mercaptoethonal | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-06-30 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of C-As lyase with mercaptoethonal
To Be Published
|
|
5C8W
 
 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MALONIC ACID, SODIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
5D4F
 
 | | Crystal structure of C-As lyase with Fe(III) | | Descriptor: | CHLORIDE ION, FE (III) ION, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-08-07 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of C-As lyase with mercaptoethonal
To Be Published
|
|
5DFG
 
 | |
5DRH
 
 | | Crystal structure of apo C-As lyase | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase, SODIUM ION | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of apo C-As lyase
To Be Published
|
|
5EVJ
 
 | | X-ray crystal structure of CrArsM, an arsenic (III) S-adenosylmethionine methyltransferase from Chlamydomonas reinhardtii | | Descriptor: | Arsenite methyltransferase, SODIUM ION | | Authors: | Packianathan, C, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CrArsM, an arsenic (III) S-adenosylmethionine methyltransferase from Chlamydomonas reinhardtii
To Be Published
|
|
8ERW
 
 | | Precisely patterned nanofibers made from extendable protein multiplexes | | Descriptor: | C2HR4_8r | | Authors: | Bick, M.J, Parmeggiani, F, Bethel, N.P, Sankaran, B, Baker, D. | | Deposit date: | 2022-10-12 | | Release date: | 2023-08-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Precisely patterned nanofibres made from extendable protein multiplexes.
Nat.Chem., 15, 2023
|
|
8CTF
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8G0W
 
 | |
8DDE
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-2-(1H-tetrazol-5-yl) yridine-3-ol | | Descriptor: | (2M)-6-bromo-3-hydroxy-2-(1H-tetrazol-5-yl)pyridin-4(1H)-one, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DJY
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-2-(4,5-dihydro-1H-imidazol-2-yl)-3-hydroxypyridin-4(1H)-one | | Descriptor: | (2M)-6-bromo-2-(4,5-dihydro-1H-imidazol-2-yl)-3-hydroxypyridin-4(1H)-one, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DDB
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-3-hydroxypicolinic acid | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DJV
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide | | Descriptor: | 6-bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DAL
 
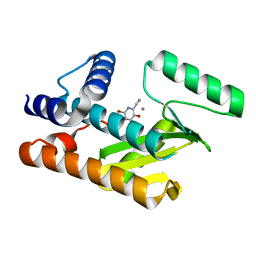 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-3-hydroxypicolinonitrile | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carbonitrile, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DHN
 
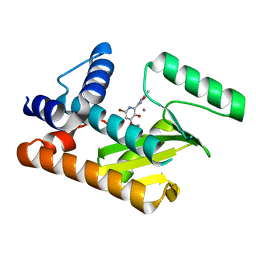 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-3-hydroxy-2-(5-methyl-1,2,4-oxadiazol-3-yl)pyridin-4(1H)-one | | Descriptor: | (2P)-6-bromo-3-hydroxy-2-(5-methyl-1,2,4-oxadiazol-3-yl)pyridin-4(1H)-one, MANGANESE (II) ION, PA endonuclease | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-27 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
4P3I
 
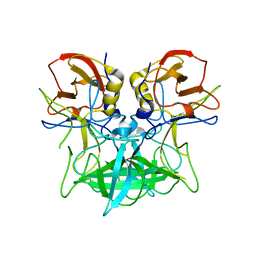 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | Descriptor: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6941 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P12
 
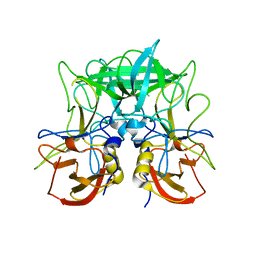 | | Native Structure of the P domain from a GI.7 Norovirus variant. | | Descriptor: | Major capsid protein | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-24 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6011 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P25
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeY HBGA. | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4991 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P26
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with A-type 2 HBGA | | Descriptor: | P domain of VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P2N
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeX HBGA | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4PM8
 
 | | Crystal structure of CTX-M-14 S70G:S237A beta-lactamase at 1.17 Angstroms resolution | | Descriptor: | Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.169 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4P1V
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with H-type 2 HBGA | | Descriptor: | P domain of VPI, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5497 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4PMA
 
 | | Crystal structure of CTX-M-14 S70G:S237A:R276A beta-lactamase at 1.39 Angstroms resolution | | Descriptor: | Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Cardenas, A.M, Adamski, C.J, Brown, N.G, Horton, L.B, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PM5
 
 | | Crystal structure of CTX-M-14 S70G beta-lactamase in complex with cefotaxime at 1.26 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
