5EQ9
 
 | |
6CD1
 
 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3), complexes with reaction intermediates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCINE, ... | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
6CZL
 
 | |
6CCZ
 
 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3) soaked with selenourea | | Descriptor: | ACETATE ION, FORMIC ACID, Serine hydroxymethyltransferase, ... | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
4Q0K
 
 | | Crystal Structure of Phytohormone Binding Protein from Medicago truncatula in complex with gibberellic acid (GA3) | | Descriptor: | GIBBERELLIN A3, GLYCEROL, PHYTOHORMONE BINDING PROTEIN MTPHBP | | Authors: | Ciesielska, A, Barciszewski, J, Ruszkowski, M, Jaskolski, M, Sikorski, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Specific binding of gibberellic acid by Cytokinin-Specific Binding Proteins: a new aspect of plant hormone-binding proteins with the PR-10 fold.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6MT1
 
 | |
6MT2
 
 | |
7QX8
 
 | | Crystal structure of serine hydroxymethyltransferase, isoform 7 from Arabidopsis thaliana (SHM7) | | Descriptor: | Serine hydroxymethyltransferase 7 | | Authors: | Ruszkowski, M, Grzechowiak, M, Sekula, B. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
7QPE
 
 | | Crystal structure of serine hydroxymethyltransferase, isoform 6 from Arabidopsis thaliana (SHM6) | | Descriptor: | NITRATE ION, Serine hydroxymethyltransferase 6 | | Authors: | Ruszkowski, M, Grzechowiak, M, Sekula, B. | | Deposit date: | 2022-01-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
7Q00
 
 | | Crystal structure of serine hydroxymethyltransferase, isoform 4 from Arabidopsis thaliana (SHM4) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Serine hydroxymethyltransferase 4 | | Authors: | Ruszkowski, M, Sekula, B. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
7PZZ
 
 | | Crystal structure of serine hydroxymethyltransferase, isoform 2 from Arabidopsis thaliana (SHM2) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Ruszkowski, M, Sekula, B. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
6YEH
 
 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in apo form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glutamate dehydrogenase 1, POTASSIUM ION | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
6YEI
 
 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in complex with NAD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
7OJ5
 
 | |
7OUO
 
 | | Crystal structure of RNA duplex [UCGUGCGA]2 in complex with Ba2+ cation | | Descriptor: | BARIUM ION, RNA (5'-R(*UP*CP*GP*UP*GP*CP*GP*A)-3') | | Authors: | Ruszkowski, M, Mao, S, Zheng, Y.Y, Ruszkowska, A, Sheng, J. | | Deposit date: | 2021-06-12 | | Release date: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Structural Insights Into the 5'UG/3'GU Wobble Tandem in Complex With Ba 2+ Cation.
Front Mol Biosci, 8, 2021
|
|
6YMF
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-Serine external aldimine state | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Serine hydroxymethyltransferase, ... | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YME
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-internal aldimine state | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YMD
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the covalent complex with malonate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MALONATE ION, ... | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6Z18
 
 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; R32 form | | Descriptor: | RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG | | Authors: | Ruszkowski, M, Sekula, B, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
8OWN
 
 | |
8OWM
 
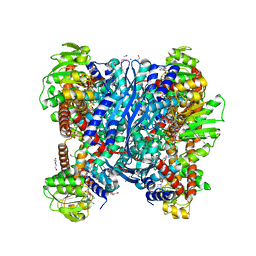 | | Crystal structure of glutamate dehydrogenase 2 from Arabidopsis thaliana binding Ca, NAD and 2,2-dihydroxyglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2,2-bis(oxidanyl)pentanedioic acid, CALCIUM ION, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional studies of Arabidopsis thaliana glutamate dehydrogenase isoform 2 demonstrate enzyme dynamics and identify its calcium binding site.
Plant Physiol Biochem., 201, 2023
|
|
8QI7
 
 | |
7Z0R
 
 | | AtWRKY18 DNA-binding domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, NITRATE ION, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
7Z0U
 
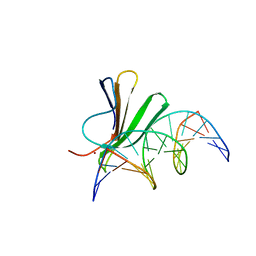 | | Crystal structure of AtWRKY18 DNA-binding domain in complex with W-box DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*GP*GP*CP*G)-3'), WRKY transcription factor 18, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
8RUF
 
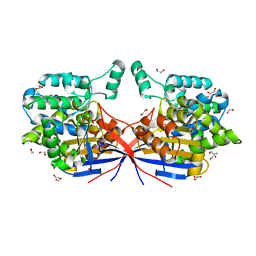 | | Crystal structure of Rhizobium etli L-asparaginase ReAV D187A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV
Front Chem, 2024
|
|
