4PQI
 
 | | Crystal structure of glutathione transferase lambda3 from Populus trichocarpa | | Descriptor: | CALCIUM ION, GLUTATHIONE, In2-1 family protein, ... | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
4PQH
 
 | | Crystal structure of glutathione transferase lambda1 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, SODIUM ION, glutathione transferase lambda1 | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
5NYM
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in reduced state | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYO
 
 | | Crystal structure of an atypical poplar thioredoxin-like2.1 variant in dimeric form | | Descriptor: | SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYK
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in oxidized state | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYN
 
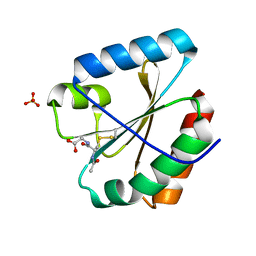 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in complex with gluathione | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYL
 
 | |
7NCW
 
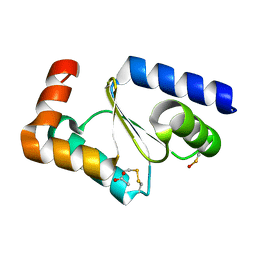 | |
7NCV
 
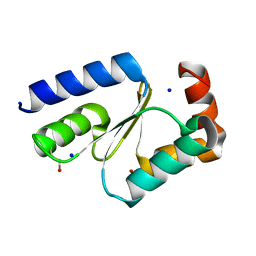 | |
6SGO
 
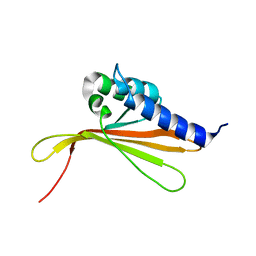 | | NMR structure of MLP124017 | | Descriptor: | Secreted protein | | Authors: | Barthe, P, de Guillen, K, Padilla, A, Hecker, A. | | Deposit date: | 2019-08-05 | | Release date: | 2019-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds.
Sci Rep, 9, 2019
|
|
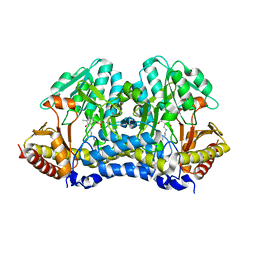
4Q76
 
 | |
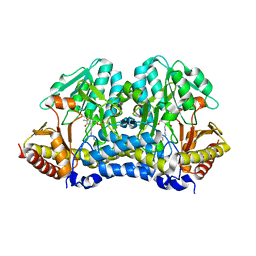
4Q75
 
 | |
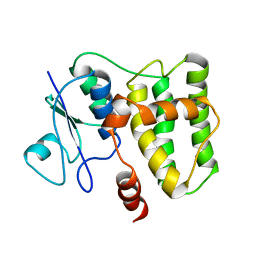
2N5F
 
 | |
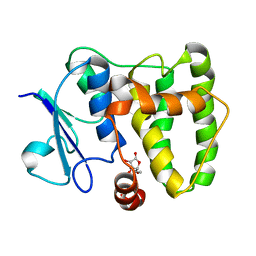
5MYE
 
 | |
5N9U
 
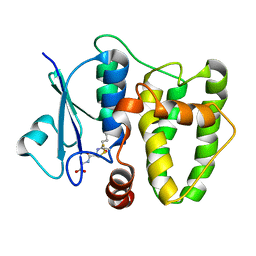 | |
5NFL
 
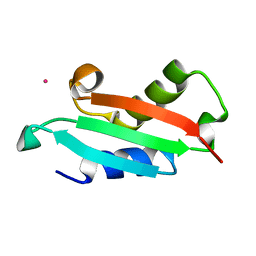 | |
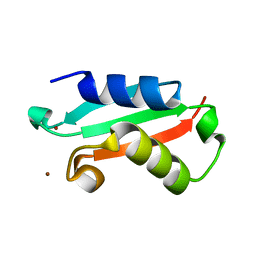
5NFK
 
 | |
5NFM
 
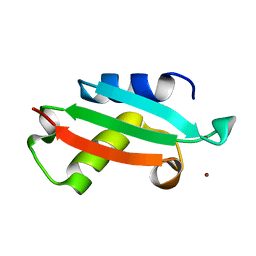 | |
4PUH
 
 | |
4PUI
 
 | |
4PUG
 
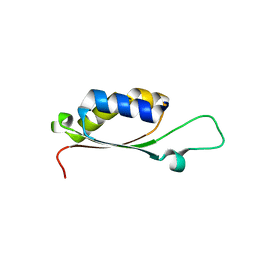 | | BolA1 from Arabidopsis thaliana | | Descriptor: | BolA like protein | | Authors: | Roret, T, Didierjean, C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural and Spectroscopic Insights into BolA-Glutaredoxin Complexes.
J.Biol.Chem., 289, 2014
|
|
8A0Q
 
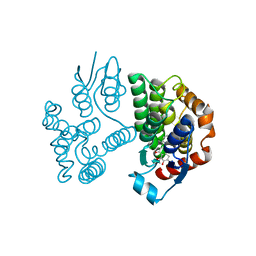 | |
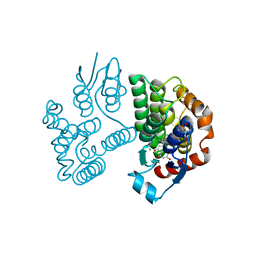
8A0R
 
 | |
8A0P
 
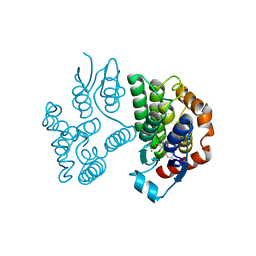 | | Crystal structure of poplar glutathione transferase U20 in complex with morin | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, CHLORIDE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
7ZS3
 
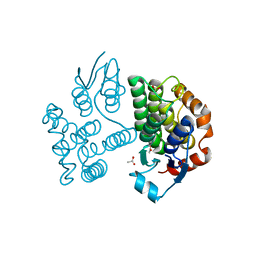 | | Crystal structure of poplar glutathione transferase U19 | | Descriptor: | ACETATE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
