1VA5
 
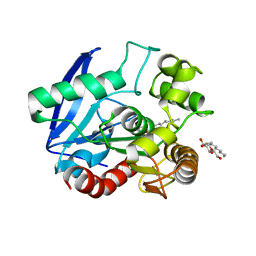 | | Antigen 85C with octylthioglucoside in active site | | Descriptor: | Antigen 85-C, octyl 1-thio-beta-D-glucopyranoside | | Authors: | Ronning, D.R, Vissa, V, Besra, G.S, Belisle, J.T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-11 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mycobacterium tuberculosis Antigen 85A and 85C Structures Confirm Binding Orientation and Conserved Substrate Specificity
J.Biol.Chem., 279, 2004
|
|
1T0F
 
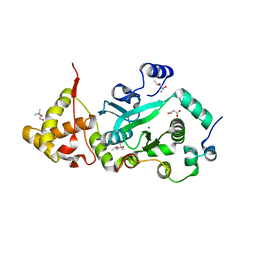 | | Crystal Structure of the TnsA/TnsC(504-555) complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MALONIC ACID, ... | | Authors: | Ronning, D.R, Li, Y, Perez, Z.N, Ross, P.D, Hickman, A.B, Craig, N.L, Dyda, F. | | Deposit date: | 2004-04-08 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA.
Embo J., 23, 2004
|
|
1SFR
 
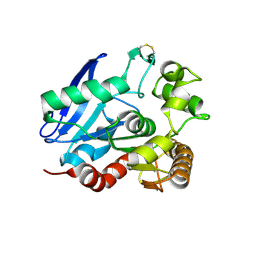 | | Crystal Structure of the Mycobacterium tuberculosis Antigen 85A Protein | | Descriptor: | Antigen 85-A | | Authors: | Ronning, D.R, Vissa, V, Besra, G.S, Belisle, J.T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-20 | | Release date: | 2004-07-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mycobacterium tuberculosis Antigen 85A and 85C Structures Confirm Binding Orientation and Conserved Substrate Specificity
J.Biol.Chem., 279, 2004
|
|
2A6M
 
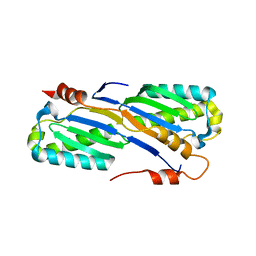 | | Crystal Structure of the ISHp608 Transposase | | Descriptor: | ISHp608 transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
2A6O
 
 | | Crystal Structure of the ISHp608 Transposase in Complex with Stem-loop DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*TP*AP*GP*CP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*GP*A)-3', ISHp608 Transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
3NM6
 
 | | Helicobacter pylori MTAN complexed with adenine and tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, ... | | Authors: | Ronning, D.R, Iacopelli, N.M. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
Protein Sci., 19, 2010
|
|
3NM5
 
 | | Helicobacter pylori MTAN complexed with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Ronning, D.R, Iacopelli, N.M. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
Protein Sci., 19, 2010
|
|
3NM4
 
 | | Helicobacter pylori MTAN | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MTA/SAH nucleosidase | | Authors: | Ronning, D.R, Iacopelli, N.M. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
Protein Sci., 19, 2010
|
|
1DQY
 
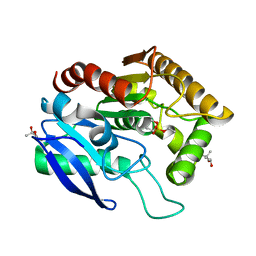 | | CRYSTAL STRUCTURE OF ANTIGEN 85C FROM MYCOBACTERIUM TUBERCULOSIS WITH DIETHYL PHOSPHATE INHIBITOR | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DIETHYL PHOSPHONATE, PROTEIN (ANTIGEN 85-C) | | Authors: | Ronning, D.R, Klabunde, T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-01-05 | | Release date: | 2000-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines.
Nat.Struct.Biol., 7, 2000
|
|
1DQZ
 
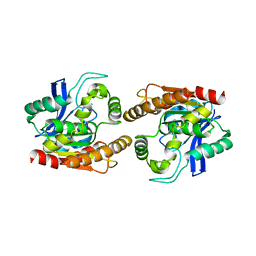 | |
4U2Z
 
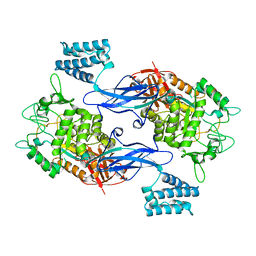 | | X-ray crystal structure of an Sco GlgEI-V279S/1,2,2-trifluromaltose complex | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-2-deoxy-2,2-difluoro-alpha-D-arabino-hexopyranosyl fluoride | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis of 2-deoxy-2,2-difluoro-alpha-maltosyl fluoride and its X-ray structure in complex with Streptomyces coelicolor GlgEI-V279S.
Org.Biomol.Chem., 13, 2015
|
|
4U2Y
 
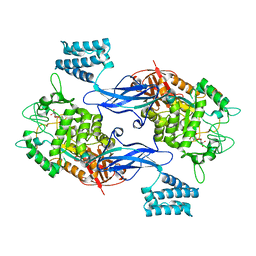 | | Sco GlgEI-V279S in Complex with Reaction Intermediate Azasugar | | Descriptor: | (2R,3R,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)pyrrolidin-3-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1 | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U31
 
 | | Sco GlgEI-V279S in Complex with maltose-C-phosphonate | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, CITRIC ACID, ... | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U33
 
 | | Structure of Mtb GlgE bound to maltose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4P54
 
 | |
4U3C
 
 | | Docking Site of Maltohexaose in the Mtb GlgE | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-19 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
5JPC
 
 | | Joint X-ray/neutron structure of MTAN complex with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, Aminodeoxyfutalosine nucleosidase | | Authors: | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8D6K
 
 | | Sco GlgEI-V279S in complex with cyclohexyl carbasugar | | Descriptor: | (1R,4S,5S,6R)-4-(cyclohexylamino)-5,6-dihydroxy-2-(hydroxymethyl)cyclohex-2-en-1-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, DI(HYDROXYETHYL)ETHER | | Authors: | Jayasinghe, T.J, Ronning, D.R. | | Deposit date: | 2022-06-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Synthesis of C 7 /C 8 -cyclitols and C 7 N-aminocyclitols from maltose and X-ray crystal structure of Streptomyces coelicolor GlgEI V279S in a complex with an amylostatin GXG-like derivative.
Front Chem, 10, 2022
|
|
3HRH
 
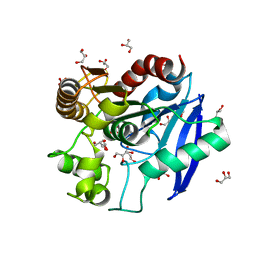 | | Crystal Structure of Antigen 85C and Glycerol | | Descriptor: | Antigen 85-C, GLYCEROL | | Authors: | Boucau, J, Sanki, A.K, Umesiri, F.E, Sucheck, S.J, Ronning, D.R. | | Deposit date: | 2009-06-09 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis and biological evaluation of sugar-derived esters, alpha-ketoesters and alpha-ketoamides as inhibitors for Mycobacterium tuberculosis antigen 85C.
Mol Biosyst, 5, 2009
|
|
1RZ9
 
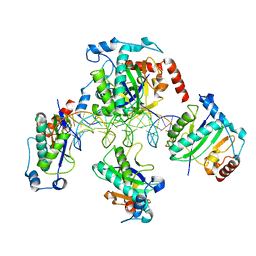 | | Crystal Structure of AAV Rep complexed with the Rep-binding sequence | | Descriptor: | 26-MER, Rep protein | | Authors: | Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M, Dyda, F. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces.
Mol.Cell, 13, 2004
|
|
8UZH
 
 | |
8F2L
 
 | |
8F5V
 
 | |
9DRK
 
 | | Crystal structure of Mycobacterium tuberculosis biotin protein ligase in complex with Bio-1 | | Descriptor: | 5'-deoxy-5'-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}sulfamamido)adenosine, Biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | McCue, W.M, Jayasinghe, Y.P, Aldrich, C.C, Ronning, D.R. | | Deposit date: | 2024-09-25 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Metabolically Stable Adenylation Inhibitors of Biotin Protein Ligase as Antibacterial Agents.
J.Med.Chem., 68, 2025
|
|
9DRN
 
 | | Crystal structure of Mycobacterium tuberculosis biotin protein ligase in complex with Bio-4 | | Descriptor: | 5'-deoxy-5'-(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)adenosine, Biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | McCue, W.M, Jayasinghe, Y.P, Aldrich, C.C, Ronning, D.R. | | Deposit date: | 2024-09-25 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Metabolically Stable Adenylation Inhibitors of Biotin Protein Ligase as Antibacterial Agents.
J.Med.Chem., 68, 2025
|
|
