5G5H
 
 | | Escherichia coli Periplasmic Aldehyde Oxidase R440H mutant | | Descriptor: | ACETATE ION, Aldehyde oxidoreductase FAD-binding subunit PaoB, Aldehyde oxidoreductase iron-sulfur-binding subunit PaoA, ... | | Authors: | Correia, M.A.S, Otrelo-Cardoso, A.R, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Escherichia Coli Periplasmic Aldehyde Oxidoreductase is an Exceptional Member of the Xanthine Oxidase Family of Molybdoenzymes.
Acs Chem.Biol., 11, 2016
|
|
2V3V
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-22 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
7OPN
 
 | | Human Aldehyde Oxidase SNP R1231H in complex with Raloxifene | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Aldehyde oxidase, DIMETHYL SULFOXIDE, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
7ORC
 
 | | Human Aldehyde Oxidase in complex with Raloxifene | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-05 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
2V45
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-27 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
3KAQ
 
 | | Flavodoxin from D. desulfuricans (semireduced form) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Romero, A, Caldeira, J, LeGall, J, Moura, I, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2009-10-19 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states.
Eur.J.Biochem., 239, 1996
|
|
3ZYV
 
 | | Crystal structure of the mouse liver Aldehyde Oxidase 3 (mAOX3) | | Descriptor: | AOX3, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Trincao, J, Coelho, C, Mahro, M, Rodrigues, D, Terao, M, Garattini, E, Leimkuehler, S, Romao, M.J. | | Deposit date: | 2011-08-27 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | The First Mammalian Aldehyde Oxidase Crystal Structure: Insights Into Substrate Specificity.
J.Biol.Chem., 287, 2012
|
|
3KAP
 
 | | Flavodoxin from Desulfovibrio desulfuricans ATCC 27774 (oxidized form) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Romero, A, Caldeira, J, LeGall, J, Moura, I, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2009-10-19 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states.
Eur.J.Biochem., 239, 1996
|
|
3L0P
 
 | | Crystal structures of Iron containing Adenylate kinase from Desulfovibrio gigas | | Descriptor: | Adenylate kinase, FE (III) ION, GLYCEROL | | Authors: | Mukhopadhyay, A, Trincao, J, Romao, M.J. | | Deposit date: | 2009-12-10 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the zinc-, cobalt-, and iron-containing adenylate kinase from Desulfovibrio gigas: a novel metal-containing adenylate kinase from Gram-negative bacteria
J.Biol.Inorg.Chem., 16, 2011
|
|
1RZ5
 
 | | Di-haem Cytochrome c Peroxidase, Form OUT | | Descriptor: | CALCIUM ION, Cytochrome c peroxidase, HEME C | | Authors: | Dias, J.M, Alves, T, Bonifacio, C, Pereira, A.S, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617.
Structure, 12, 2004
|
|
6SDV
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Formate reduced form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
6SDR
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Oxidized form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, alpha subunit, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
1RZ6
 
 | | Di-haem Cytochrome c Peroxidase, Form IN | | Descriptor: | CITRIC ACID, Cytochrome c peroxidase, HEME C | | Authors: | Dias, J.M, Alves, T, Bonifacio, C, Pereira, A, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617.
Structure, 12, 2004
|
|
2JIR
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CYANIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2JIP
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2JIO
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2JIQ
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2JIM
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2C4X
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-10-25 | | Release date: | 2005-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
2XJW
 
 | | Lysozyme-CO releasing molecule adduct | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, LYSOZYME C, ... | | Authors: | Santos-Silva, T, Mukhopadhyay, A, Romao, M.J. | | Deposit date: | 2010-07-06 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Corm-3 Reactivity Toward Proteins: The Crystal Structure of a Ru(II) Dicarbonyl-Lysozyme Complex.
J.Am.Chem.Soc., 133, 2011
|
|
1JAF
 
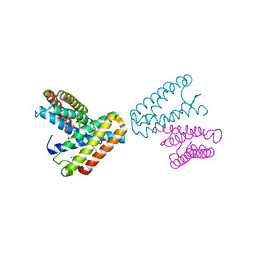 | | CRYSTAL STRUCTURE OF CYTOCHROME C' FROM RHODOCYCLUS GELATINOSUS AT 2.5 ANGSTOMS RESOLUTION | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Archer, M, Banci, L, Dikaya, E, Romao, M.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Cytochrome C' from Rhodocyclus Gelatinosus and Comparison with Other Cytochromes C'
J.Biol.Inorg.Chem., 2, 1997
|
|
1GVZ
 
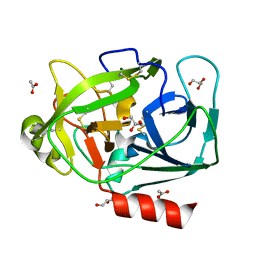 | | Prostate Specific Antigen (PSA) from stallion seminal plasma | | Descriptor: | ACETATE ION, GLYCEROL, KALLIKREIN-1E2 | | Authors: | Carvalho, A.L, Sanz, L, Barettino, D, Romero, A, Calvete, J.J, Romao, M.J. | | Deposit date: | 2002-02-28 | | Release date: | 2002-09-12 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of a Prostate Kallikrein Isolated from Stallion Seminal Plasma: A Homologue of Human Psa
J.Mol.Biol., 322, 2002
|
|
8CCQ
 
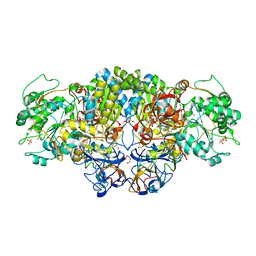 | | Crystal structure of arsenite oxidase from Pseudorhizobium banfieldiae str. NT-26 (NT-26 Aio) bound to antimony trioxide | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AroA, ... | | Authors: | Engrola, F, Santos-Silva, T, Romao, M.J, Correia, M.A.S. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Arsenite oxidase in complex with antimonite and arsenite oxyanions: Insights into the catalytic mechanism.
J.Biol.Chem., 299, 2023
|
|
8CFF
 
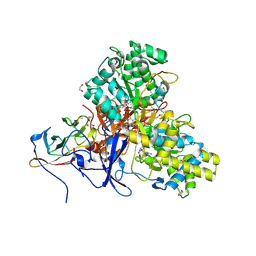 | | Crystal structure of arsenite oxidase from Alcaligenes faecalis (Af Aio) bound to arsenite | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENITE, ... | | Authors: | Engrola, F, Correia, M.A.S, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Arsenite oxidase in complex with antimonite and arsenite oxyanions: Insights into the catalytic mechanism.
J.Biol.Chem., 299, 2023
|
|
