5KYM
 
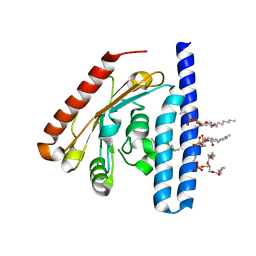 | | Crystal Structure of the 1-acyl-sn-glycerophosphate (LPA) acyltransferase, PlsC, from Thermotoga maritima | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, 1-acyl-sn-glycerol-3-phosphate acyltransferase, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Robertson, R.M, Yao, J, Gajewski, S, Kumar, G, Martin, E.W, Rock, C.O, White, S.W. | | Deposit date: | 2016-07-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A two-helix motif positions the lysophosphatidic acid acyltransferase active site for catalysis within the membrane bilayer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5JLT
 
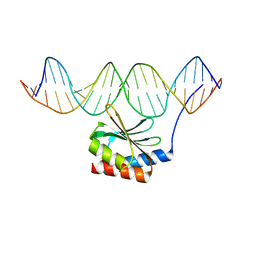 | | The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*TP*GP*CP*TP*TP*AP*AP*TP*AP*AP*TP*CP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*GP*AP*TP*TP*AP*TP*TP*AP*AP*GP*CP*AP*AP*AP*GP*CP*TP*TP*C)-3'), Middle transcription regulatory protein motA | | Authors: | Cuypers, M.G, Robertson, R.M, Knipling, L, Hinton, D.M, White, S.W. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA.
Nucleic Acids Res., 46, 2018
|
|
4Q9N
 
 | | Crystal structure of Chlamydia trachomatis enoyl-ACP reductase (FabI) in complex with NADH and AFN-1252 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | Authors: | Yao, J, Abdelrahman, Y, Robertson, R.M, Cox, J.V, Belland, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Type II Fatty Acid Synthesis Is Essential for the Replication of Chlamydia trachomatis.
J.Biol.Chem., 289, 2014
|
|
