7VYV
 
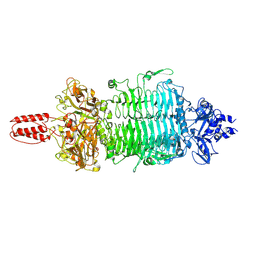 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Depo32, a depolymerase of Klebsiella phage, efficiently controls infection caused by Klebsiella pneumoniae with K2 serotype CPS
To Be Published
|
|
7VZ3
 
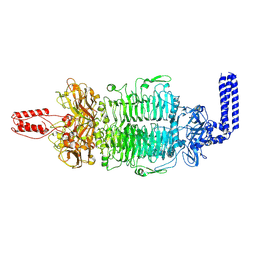 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Depo32, a depolymerase of Klebsiella phage, efficiently controls infection caused by Klebsiella pneumoniae with K2 serotype CPS
To Be Published
|
|
6IUG
 
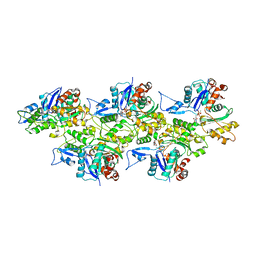 | | Cryo-EM structure of the plant actin filaments from Zea mays pollen | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, pollen F-actin | | Authors: | Ren, Z.H, Zhang, Y, Zhang, Y, He, Y.Q, Du, P.Z, Wang, Z.X, Sun, F, Ren, H.Y. | | Deposit date: | 2018-11-28 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of Actin Filaments fromZea maysPollen.
Plant Cell, 31, 2019
|
|
1AX9
 
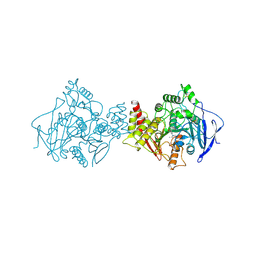 | | ACETYLCHOLINESTERASE COMPLEXED WITH EDROPHONIUM, LAUE DATA | | Descriptor: | ACETYLCHOLINESTERASE, EDROPHONIUM ION | | Authors: | Raves, M.L, Ravelli, R.B.G, Sussman, J.L, Harel, M, Silman, I. | | Deposit date: | 1997-11-03 | | Release date: | 1998-02-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Static Laue diffraction studies on acetylcholinesterase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2ACK
 
 | | ACETYLCHOLINESTERASE COMPLEXED WITH EDROPHONIUM, MONOCHROMATIC DATA | | Descriptor: | ACETYLCHOLINESTERASE, EDROPHONIUM ION | | Authors: | Raves, M.L, Sussman, J.L, Harel, M, Silman, I. | | Deposit date: | 1997-10-29 | | Release date: | 1998-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Static Laue diffraction studies on acetylcholinesterase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6PH2
 
 | | Complete LOV domain from the LOV-HK sensory protein from Brucella abortus (mutant C69S, construct 15-155) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PH4
 
 | | Full length LOV-PAS-HK construct from the LOV-HK sensory protein from Brucella abortus (light-adapted, construct 15-489) | | Descriptor: | Blue-light-activated histidine kinase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PH3
 
 | | LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (dark-adapted, construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PPS
 
 | | A blue light illuminated LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Fernandez, I, Shin, H, Gunawardana, S, Otero, L.H, Cerutti, M.L, Yang, X, Klinke, S, Goldbaum, F.A. | | Deposit date: | 2019-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6ITP
 
 | | Crystal structure of cortisol complexed with its nanobody at pH 3.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
6ITQ
 
 | | Crystal structure of cortisol complexed with its nanobody at pH 10.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, SULFATE ION, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
6JYT
 
 | |
5K3E
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Glycolate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3B
 
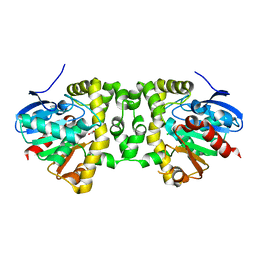 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Chloroacetate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, chloroacetic acid | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3A
 
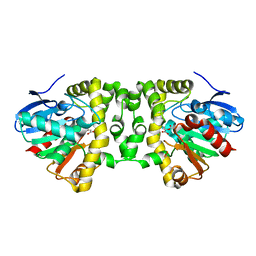 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Both Protomers Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3D
 
 | |
5K3C
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/5-Fluorotryptophan | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3F
 
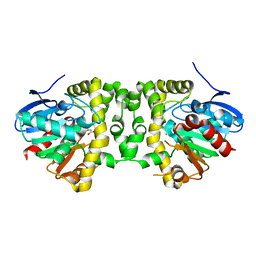 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Single Protomer Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5KCM
 
 | | Crystal structure of iron-sulfur cluster containing photolyase PhrB mutant I51W | | Descriptor: | (6-4) photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, X, Bowatte, K, Zhang, F, Lamparter, T. | | Deposit date: | 2016-06-06 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
5LFA
 
 | | Crystal structure of iron-sulfur cluster containing bacterial (6-4) photolyase PhrB - Y424F mutant with impaired DNA repair activity | | Descriptor: | (6-4) photolyase, 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kwiatkowski, D, Zhang, F, Krauss, N, Lamparter, T, Scheerer, P. | | Deposit date: | 2016-06-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
8GBO
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-AIB-ALA-AIB-LEU-CYS-GLN-AIB-LEU, NICOTINIC ACID | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-02-26 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GBA
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-(AIB)-ALA-SER-LEU-ALA-CYS-(AIB)-LEU, NICOTINIC ACID | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-02-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GBM
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETATE ION, LEAD (II) ION, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-02-26 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GD6
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, NICOTINIC ACID, NIO-LEU-AIB-ALA-AIB-LEU-AIB-CYS-AIB-LEU-BPH | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GBI
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETONITRILE, LEU-(AIB)-ALA-CYS-LEU-CYS-CYS-(AIB)-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-02-26 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
