6EV1
 
 | | Crystal structure of antibody against schizophyllan | | Descriptor: | Heavy chain, Light chain | | Authors: | Sung, K.H, Josewski, J, Dubel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
6EV2
 
 | | Crystal structure of antibody against schizophyllan in complex with laminarihexaose | | Descriptor: | Heavy chain, Light chain, beta-D-glucopyranose, ... | | Authors: | Sung, K.H, Josewski, J, Duebel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
4KUK
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
4KUO
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Photoexcited state) | | Descriptor: | RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
4FBM
 
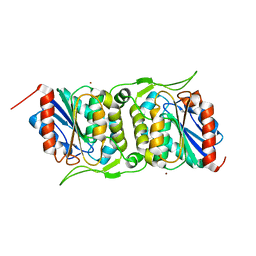 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
4FBL
 
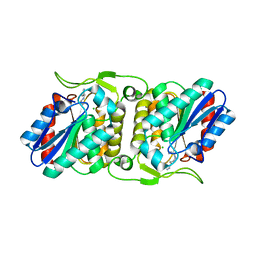 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
2FWG
 
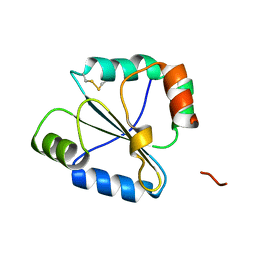 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (photoreduced form) | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWH
 
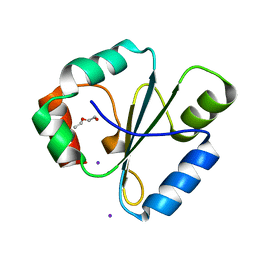 | | atomic resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form at pH7) | | Descriptor: | DI(HYDROXYETHYL)ETHER, IODIDE ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWF
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form) | | Descriptor: | IODIDE ION, SODIUM ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWE
 
 | | crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (oxidized form) | | Descriptor: | IODIDE ION, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2YOM
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Lecher, J, Hartmann, R, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
2YON
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Hartmann, R, Lecher, J, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
1VRS
 
 | | Crystal structure of the disulfide-linked complex between the N-terminal and C-terminal domain of the electron transfer catalyst DsbD | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Rozhkova, A, Stirnimann, C.U, Frei, P, Grauschopf, U, Brunisholz, R, Gruetter, M.G, Capitani, G, Glockshuber, R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD
Embo J., 23, 2004
|
|
3SW1
 
 | | Structure of a full-length bacterial LOV protein | | Descriptor: | FLAVIN MONONUCLEOTIDE, Sensory box protein | | Authors: | Granzin, J, Batra-Safferling, R, Jaeger, K.-E, Drepper, T, Krauss, U. | | Deposit date: | 2011-07-13 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural Basis for the Slow Dark Recovery of a Full-Length LOV Protein from Pseudomonas putida.
J.Mol.Biol., 417, 2012
|
|
1Z5Y
 
 | | Crystal Structure Of The Disulfide-Linked Complex Between The N-Terminal Domain Of The Electron Transfer Catalyst DsbD and The Cytochrome c Biogenesis Protein CcmG | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thiol:disulfide interchange protein dsbD, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Gruetter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis and Kinetics of DsbD-Dependent Cytochrome c Maturation
STRUCTURE, 13, 2005
|
|
8A33
 
 | |
2WCD
 
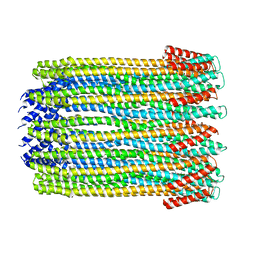 | | Crystal structure of the assembled cytolysin A pore | | Descriptor: | ETHYL MERCURY ION, HEMOLYSIN E, CHROMOSOMAL | | Authors: | Mueller, M, Grauschopf, U, Maier, T, Glockshuber, R, Ban, N. | | Deposit date: | 2009-03-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | The Structure of a Cytolytic Alpha-Helical Toxin Pore Reveals its Assembly Mechanism
Nature, 459, 2009
|
|
8T6F
 
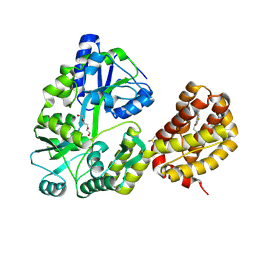 | | Crystal structure of human MBP-Myeloid cell leukemia 1 (Mcl-1) in complex with BRD810 inhibitor | | Descriptor: | (3aM,9S,15R)-4-chloro-3-ethyl-7-{3-[(6-fluoronaphthalen-1-yl)oxy]propyl}-2-methyl-15-[2-(morpholin-4-yl)ethyl]-2,10,11,12,13,15-hexahydropyrazolo[4',3':9,10][1,6]oxazacycloundecino[8,7,6-hi]indole-8-carboxylic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Poncet-Montange, G, Lemke, C.T. | | Deposit date: | 2023-06-15 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | BRD-810 is a highly selective MCL1 inhibitor with optimized in vivo clearance and robust efficacy in solid and hematological tumor models.
Nat Cancer, 5, 2024
|
|
6YXB
 
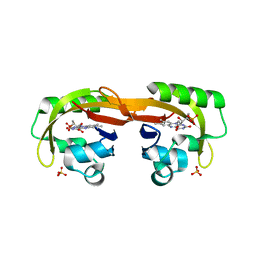 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (space group P21) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX6
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (no morpholine) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX4
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant with morpholine | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7AB7
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) I52T Q148K variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-09-06 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7ABY
 
 | | Crystal structure of iLOV-Q489K mutant | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Granzin, J, Batra-Safferling, R. | | Deposit date: | 2020-09-09 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7AB6
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) I52T variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-09-06 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
3PLK
 
 | | Bovine trypsin variant X(tripleIle227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Braeuer, U, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
