3BQF
 
 | |
8QL0
 
 | |
4YEB
 
 | | Structural characterization of a synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibronectin leucine rich transmembrane protein 3, ... | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, von Daake, S, Li, S, Lee, D, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2015-02-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
3UT5
 
 | | Tubulin-Colchicine-Ustiloxin: Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ranaivoson, F.M, Gigant, B, Knossow, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural plasticity of tubulin assembly probed by vinca-domain ligands.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6DLF
 
 | | Crystal structure of NTRI homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neurotrimin | | Authors: | Ranaivoson, F.M, Turk, L.S, Ozkan, E, Montelione, G.T, Comoletti, D. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.446 Å) | | Cite: | Structure of a heterodimer of neuronal cell surface proteins
Structure, 2019
|
|
8BJS
 
 | | Apo KIF20A[55-510] crystal structure | | Descriptor: | Kinesin-like protein KIF20A, SULFATE ION, UNK-UNK-UNK-UNK | | Authors: | Ranaivoson, F.M, Kikuti, C, Crozet, V, Sirkia, M.E, Houdusse, A. | | Deposit date: | 2022-11-06 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Nucleotide-free structures of KIF20A illuminate atypical mechanochemistry in this kinesin-6.
Open Biology, 13, 2023
|
|
2FY6
 
 | | Structure of the N-terminal domain of Neisseria meningitidis PilB | | Descriptor: | CHLORIDE ION, Peptide methionine sulfoxide reductase msrA/msrB, SULFATE ION | | Authors: | Ranaivoson, F.M, Kauffmann, B, Neiers, F, Boschi-Muller, S, Branlant, G, Favier, F. | | Deposit date: | 2006-02-07 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Structure of the N-terminal Domain of PILB from Neisseria meningitidis Reveals a Thioredoxin-fold
J.Mol.Biol., 358, 2006
|
|
6DLD
 
 | | Crystal structure of IgLON5/NEGR1 heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IgLON family member 5, Neuronal growth regulator 1, ... | | Authors: | Ranaivoson, F.M, Turk, L.S, Ozkan, E, Montelione, G.T, Comoletti, D. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a heterodimer of neuronal cell surface proteins
Structure, 2019
|
|
6DLE
 
 | | Crystal structure of IgLON5 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgLON family member 5 | | Authors: | Ranaivoson, F.M, Turk, L.S, Ozkan, E, Montelione, G.T, Comoletti, D. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.994 Å) | | Cite: | Structure of a heterodimer of neuronal cell surface proteins
Structure, 2019
|
|
4EB6
 
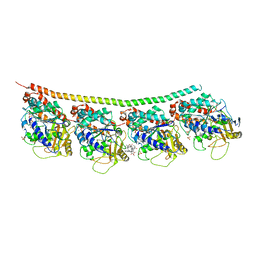 | | Tubulin-Vinblastine: Stathmin-like complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ranaivoson, F.M, Gigant, B, Knossow, M. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural plasticity of tubulin assembly probed by vinca-domain ligands.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4RMK
 
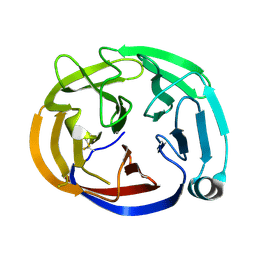 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in P65 crystal form | | Descriptor: | CALCIUM ION, Latrophilin-3 | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
4RML
 
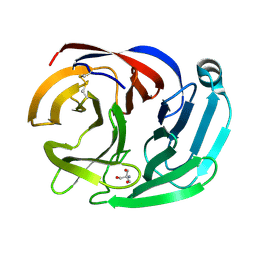 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in C2221 crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Latrophilin-3, MAGNESIUM ION | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
3BQG
 
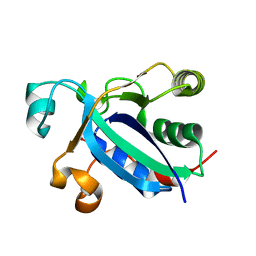 | |
3BQE
 
 | |
3BQH
 
 | |
3HCG
 
 | |
3HCI
 
 | | Structure of MsrB from Xanthomonas campestris (complex-like form) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, CALCIUM ION, Peptide methionine sulfoxide reductase, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
3HCJ
 
 | |
3HCH
 
 | | Structure of the C-terminal domain (MsrB) of Neisseria meningitidis PilB (complex with substrate) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
8F1A
 
 | | Apo KIF20A[1-565] class-1 in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF20A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Crozet, V, Ranaivoson, F.M, Houdusse, A, Sosa, H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nucleotide-free structures of KIF20A illuminate atypical mechanochemistry in this kinesin-6.
Open Biology, 13, 2023
|
|
8F18
 
 | | Apo KIF20A[1-565] class-2 in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF20A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Crozet, V, Ranaivoson, F.M, Houdusse, A, Sosa, H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nucleotide-free structures of KIF20A illuminate atypical mechanochemistry in this kinesin-6.
Open Biology, 13, 2023
|
|
5FTT
 
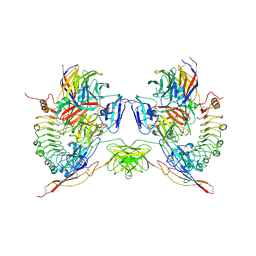 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
5FTU
 
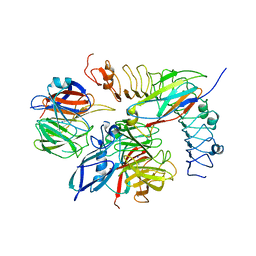 | | Tetrameric complex of Latrophilin 3, Unc5D and FLRT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.01 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
6HKC
 
 | |
