4FAU
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Li+, Mg2+ and 5'-exon | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Group IIC intron, MAGNESIUM ION, ... | | Authors: | Marcia, M, Pyle, A.M. | | Deposit date: | 2012-05-22 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Visualizing Group II Intron Catalysis through the Stages of Splicing.
Cell(Cambridge,Mass.), 151, 2012
|
|
4FAW
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+ and a hydrolyzed oligonucleotide fragment | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-R(*A*UP*UP*UP*AP*UP*UP*A)-3', Group IIC intron, ... | | Authors: | Marcia, M, Pyle, A.M. | | Deposit date: | 2012-05-22 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Visualizing Group II Intron Catalysis through the Stages of Splicing.
Cell(Cambridge,Mass.), 151, 2012
|
|
4FAR
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+ and 5'-exon | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Group IIC intron, MAGNESIUM ION, ... | | Authors: | Marcia, M, Pyle, A.M. | | Deposit date: | 2012-05-22 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Visualizing Group II Intron Catalysis through the Stages of Splicing.
Cell(Cambridge,Mass.), 151, 2012
|
|
4FAQ
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Ca2+ and 5'-exon | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Group IIC intron, ... | | Authors: | Marcia, M, Pyle, A.M. | | Deposit date: | 2012-05-22 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Visualizing Group II Intron Catalysis through the Stages of Splicing.
Cell(Cambridge,Mass.), 151, 2012
|
|
4FB0
 
 | |
7UIM
 
 | |
7UIN
 
 | |
5UC6
 
 | |
6M6R
 
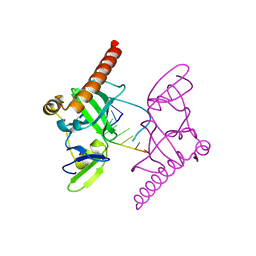 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 8-mer ssRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*CP*GP*CP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6M6Q
 
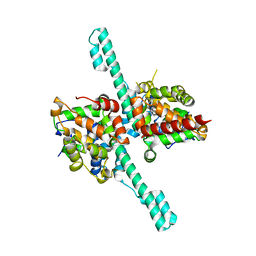 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) N-terminal domain | | Descriptor: | Dicer Related Helicase | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6M6S
 
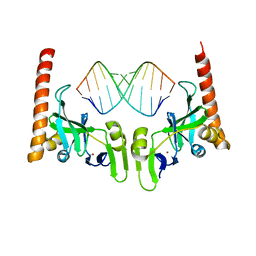 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 12-mer dsRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
3O8R
 
 | |
6Q39
 
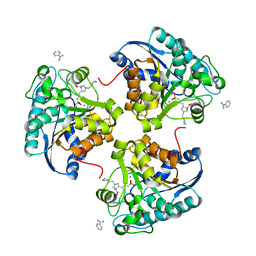 | | Complex of Arginase 2 with Example 49 | | Descriptor: | 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-1-[[(2~{S})-piperidin-2-yl]methyl]pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, mitochondrial, ... | | Authors: | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
6Q37
 
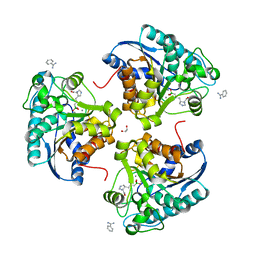 | | Complex of Arginase 2 with Example 23 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, ... | | Authors: | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
3G78
 
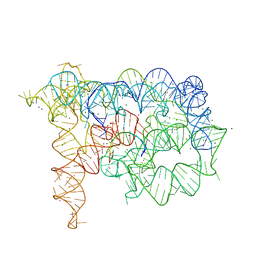 | | Insight into group II intron catalysis from revised crystal structure | | Descriptor: | Group II intron, Ligated EXON product, MAGNESIUM ION, ... | | Authors: | Wang, J. | | Deposit date: | 2009-02-09 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inclusion of weak high-resolution X-ray data for improvement of a group II intron structure.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3O8D
 
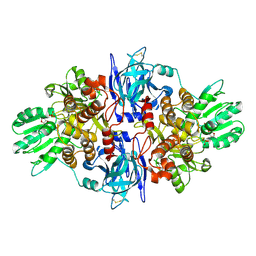 | |
3O8C
 
 | |
3O8B
 
 | |
2LPS
 
 | |
