5F75
 
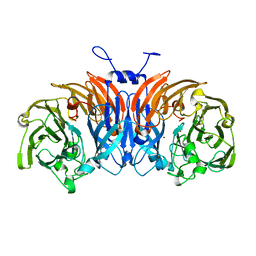 | | Thiocyanate dehydrogenase from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, Thiocyanate dehydrogenase | | Authors: | Tsallagov, S.I, Polyakov, K.M, Tikhonova, T.V, Trofimov, A.A, Shabalin, I.G, Popov, A.N, Popov, V.O. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thiocyanate dehydrogenase from Thioalkalivibrio paradoxus
To Be Published
|
|
7Q2W
 
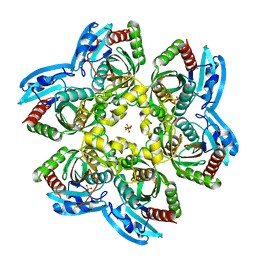 | |
3SXQ
 
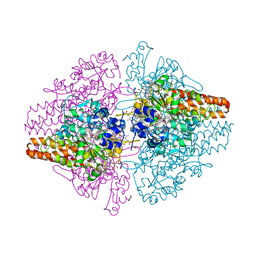 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio paradoxus | | Descriptor: | CALCIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Boyko, K.M, Popov, V.O. | | Deposit date: | 2011-07-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
3TTB
 
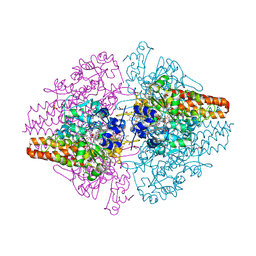 | | Structure of the Thioalkalivibrio paradoxus cytochrome c nitrite reductase in complex with sulfite | | Descriptor: | CALCIUM ION, COBALT (II) ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-09-14 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
6I3Q
 
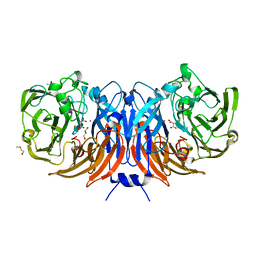 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus complex with acetate ions. | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Polyakov, K.M, Popov, A.N, Tikhkonova, T.V, Popov, V.O, Trofimov, A.A. | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4F8X
 
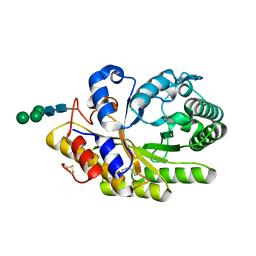 | | Penicillium canescens endo-1,4-beta-xylanase XylE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-1,4-beta-xylanase, beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Polyakov, K.M, Trofimov, A.A, Fedorova, T.V, Koroleva, O.V, Maisuradze, I.G, Chulkin, A.M, Vavilova, E.A, Benevolenskii, S.V. | | Deposit date: | 2012-05-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: |
|
|
6G50
 
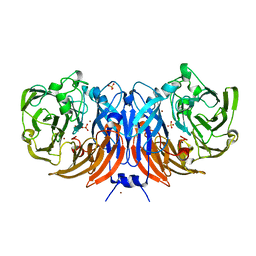 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus as isolated. | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SULFATE ION, ... | | Authors: | Polyakov, K.M, Tsallagov, S.I, Tikhkonova, T.V, Popov, V.O. | | Deposit date: | 2018-03-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SJI
 
 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus mutant with His 482 replaced by Gln | | Descriptor: | COPPER (II) ION, SULFATE ION, thiocyanate dehydrogenase | | Authors: | Polyakov, K.M, Tikhonova, T.V, Rakitina, T.V, Osipov, E, Popov, V.O. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4R2W
 
 | | X-ray structure of uridine phosphorylase from Shewanella oneidensis MR-1 in complex with uridine at 1.6 A resolution | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE, ... | | Authors: | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Veiko, V.P, Popov, V.O, Polyakov, K.P. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-syn conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis MR-1 uridine phosphorylase in the free form and in complex with uridine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1G8T
 
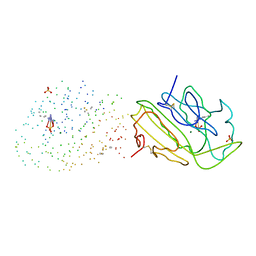 | | SM ENDONUCLEASE FROM SERATIA MARCENSCENS AT 1.1 A RESOLUTION | | Descriptor: | MAGNESIUM ION, NUCLEASE SM2 ISOFORM, SULFATE ION | | Authors: | Lunin, V.V, Perbandt, M, Betzel, C.H, Mikhailov, A.M. | | Deposit date: | 2000-11-21 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structure of the Serratia marcescens endonuclease at 1.1 A resolution and the enzyme reaction mechanism.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5EEB
 
 | | Apo form of thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 | | Descriptor: | Aldehyde dehydrogenase | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-10-22 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.038 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EKC
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Shabalin, I.G, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure of thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+
To Be Published
|
|
5EXF
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5F2C
 
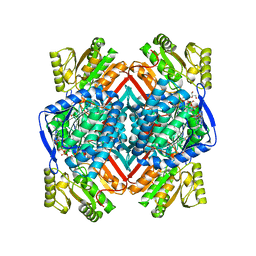 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 crystallized in microgravity (complex with NADP+) | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-12-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EUY
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
1I40
 
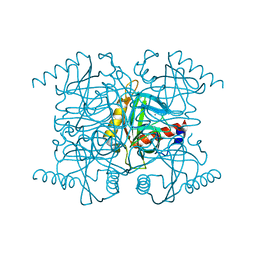 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Samygina, V.R, Popov, A.N, Lamzin, V.S, Avaeva, S.M. | | Deposit date: | 2001-02-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The structures of Escherichia coli inorganic pyrophosphatase complexed with Ca(2+) or CaPP(i) at atomic resolution and their mechanistic implications.
J.Mol.Biol., 314, 2001
|
|
1I6T
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Samygina, V.R, Popov, A.N, Lamzin, V.S, Avaeva, S.M. | | Deposit date: | 2001-03-05 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structures of Escherichia coli inorganic pyrophosphatase complexed with Ca(2+) or CaPP(i) at atomic resolution and their mechanistic implications.
J.Mol.Biol., 314, 2001
|
|
3SQR
 
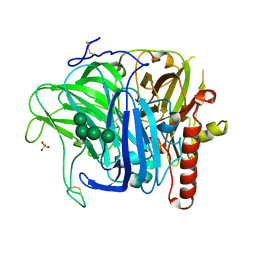 | | Crystal structure of laccase from Botrytis aclada at 1.67 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, SULFATE ION, ... | | Authors: | Osipov, E.M, Polyakov, K.M, Tikhonova, T.V, Dorovatovsky, P.V, Ludwig, R, Kittl, R, Shleev, S.V, Popov, V.O. | | Deposit date: | 2011-07-06 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3V9E
 
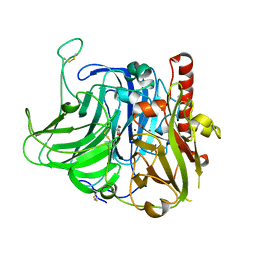 | | Structure of the L499M mutant of the laccase from B.aclada | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Osipov, E.M, Polyakov, K.M, Tikhonova, T.V, Dorovatovsky, P.V, Ludwig, R, Kittl, R, Shleev, S.V, Popov, V.O. | | Deposit date: | 2011-12-27 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1QL0
 
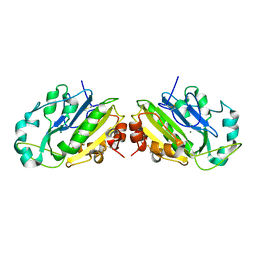 | |
1RGH
 
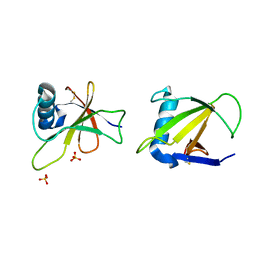 | | HYDROLASE, GUANYLORIBONUCLEASE | | Descriptor: | RIBONUCLEASE, SULFATE ION | | Authors: | Sevcik, J, Dauter, Z, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1995-06-05 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ribonuclease from Streptomyces aureofaciens at atomic resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1RGE
 
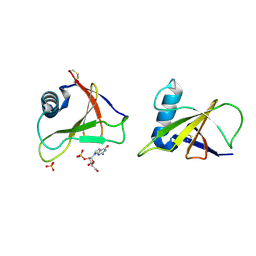 | | HYDROLASE, GUANYLORIBONUCLEASE | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE, SULFATE ION | | Authors: | Sevcik, J, Dauter, Z, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1995-06-05 | | Release date: | 1996-10-14 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ribonuclease from Streptomyces aureofaciens at atomic resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1RSN
 
 | | RIBONUCLEASE (RNASE SA) (E.C.3.1.4.8) COMPLEXED WITH EXO-2',3'-CYCLOPHOSPHOROTHIOATE | | Descriptor: | GUANOSINE-2',3'-CYCLOPHOSPHOROTHIOATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Dauter, Z, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of ribonuclease Sa with a cyclic nucleotide and a proposed model for the reaction intermediate.
Eur.J.Biochem., 216, 1993
|
|
1RGF
 
 | | HYDROLASE, GUANYLORIBONUCLEASE | | Descriptor: | RIBONUCLEASE, SULFATE ION | | Authors: | Sevcik, J, Dauter, Z, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1995-06-05 | | Release date: | 1996-10-14 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ribonuclease from Streptomyces aureofaciens at atomic resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1RGG
 
 | | HYDROLASE, GUANYLORIBONUCLEASE | | Descriptor: | RIBONUCLEASE, SULFATE ION | | Authors: | Sevcik, J, Dauter, Z, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1995-06-05 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ribonuclease from Streptomyces aureofaciens at atomic resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
