8OGI
 
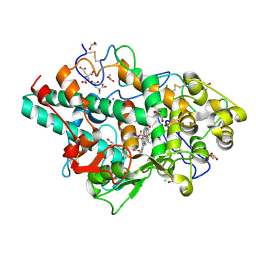 | | Structure of native human eosinophil peroxidase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pfanzagl, V, Obinger, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Posttranslational modification and heme cavity architecture of human eosinophil peroxidase-insights from first crystal structure and biochemical characterization.
J.Biol.Chem., 299, 2023
|
|
6FIY
 
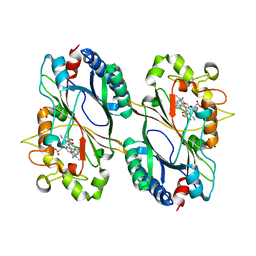 | | Crystal structure of a dye-decolorizing peroxidase D143AR232A variant from Klebsiella pneumoniae (KpDyP) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Hofbauer, S, Mlynek, G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09000432 Å) | | Cite: | Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage.
J. Biol. Chem., 293, 2018
|
|
6FKT
 
 | |
6FKS
 
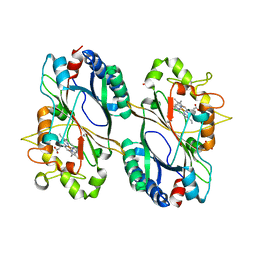 | | Crystal structure of a dye-decolorizing peroxidase from Klebsiella pneumoniae (KpDyP) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Hofbauer, S, Mlynek, G. | | Deposit date: | 2018-01-24 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.60000467 Å) | | Cite: | Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage.
J. Biol. Chem., 293, 2018
|
|
6FL2
 
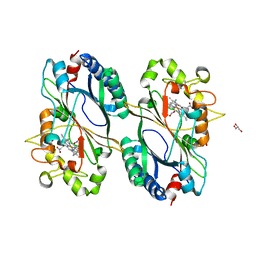 | | Crystal structure of a dye-decolorizing peroxidase D143A variant from Klebsiella pneumoniae (KpDyP) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Hofbauer, S, Mlynek, G. | | Deposit date: | 2018-01-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.270001 Å) | | Cite: | Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage.
J. Biol. Chem., 293, 2018
|
|
6RPE
 
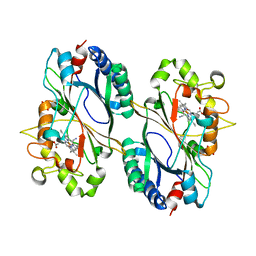 | | Structure of 5% reduced KpDyP in complex with cyanide | | Descriptor: | CYANIDE ION, GLYCEROL, Iron-dependent peroxidase, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RPD
 
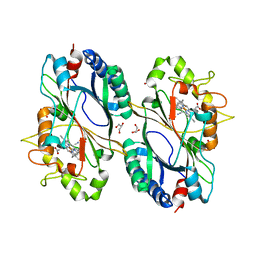 | | Structure of ferrous KpDyP in complex with cyanide | | Descriptor: | CYANIDE ION, GLYCEROL, Iron-dependent peroxidase, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RR6
 
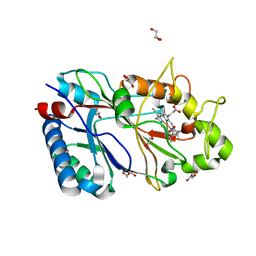 | | Structure of 100% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RQY
 
 | | Structure of % reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RR5
 
 | | Structure of 50% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RR4
 
 | | Structure of 25% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RR8
 
 | | Structure of 100% reduced KpDyP (final wedges) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RR1
 
 | | Structure of 10% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
7QZR
 
 | |
7Z53
 
 | |
6XUC
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Chlorite dismutase | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8702 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
6XUB
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with monovinyl monopropionyl deuteroheme | | Descriptor: | Chlorite dismutase, harderoheme (III) | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
6FXJ
 
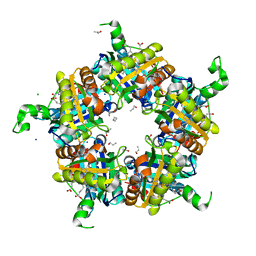 | | Structure of coproheme decarboxylase from Listeria monocytogenes in complex with iron coproporphyrin III | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, CHLORIDE ION, N-PROPANOL, ... | | Authors: | Hofbauer, S, Pfanzagl, V, Mlynek, G. | | Deposit date: | 2018-03-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Redox Cofactor Rotates during Its Stepwise Decarboxylation: Molecular Mechanism of Conversion of Coproheme to Hemeb.
Acs Catalysis, 9, 2019
|
|
6FXQ
 
 | | Structure of coproheme decarboxylase from Listeria monocytogenes during turnover | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Hofbauer, S, Pfanzagl, V, Mlynek, G, Puehringer, D. | | Deposit date: | 2018-03-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Redox Cofactor Rotates during Its Stepwise Decarboxylation: Molecular Mechanism of Conversion of Coproheme to Hemeb.
Acs Catalysis, 9, 2019
|
|
8AW7
 
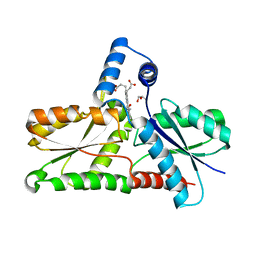 | | Structure of coproporphyrin III-LmCpfC R45L | | Descriptor: | Coproporphyrin III ferrochelatase, GLYCEROL, coproporphyrin III | | Authors: | Gabler, T, Hofbauer, S, Pfanzagl, V. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Active site architecture of coproporphyrin ferrochelatase with its physiological substrate coproporphyrin III: Propionate interactions and porphyrin core deformation.
Protein Sci., 32, 2023
|
|
7OU5
 
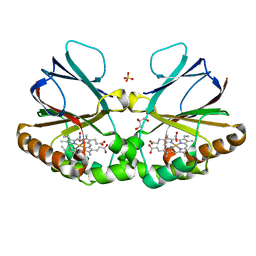 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite Dismutase, GLYCEROL, NITRITE ION, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OU7
 
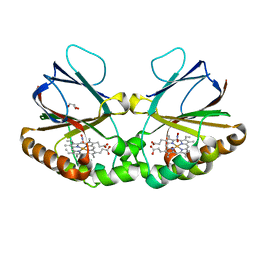 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite dismutase, GLYCEROL, NITRITE ION, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OWI
 
 | | Crystal structure of dimeric chlorite dismutase variant R127A (CCld R127A) from Cyanothece sp. PCC7425 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OU9
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74E (CCld Q74E) from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite dismutase, GLYCEROL, NITRITE ION, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OUA
 
 | | Crystal structure of dimeric chlorite dismutase variant R127K (CCld R127K) from Cyanothece sp. PCC7425 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
