3SYN
 
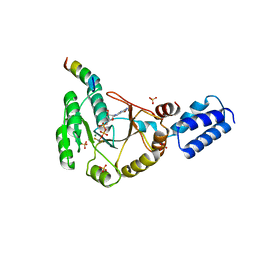 | | Crystal structure of FlhF in complex with its activator | | 分子名称: | ALUMINUM FLUORIDE, ATP-binding protein YlxH, Flagellar biosynthesis protein flhF, ... | | 著者 | Bange, G, Kuemmerer, N, Wild, K, Sinning, I. | | 登録日 | 2011-07-18 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.063 Å) | | 主引用文献 | Structural basis for the molecular evolution of SRP-GTPase activation by protein.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2PX3
 
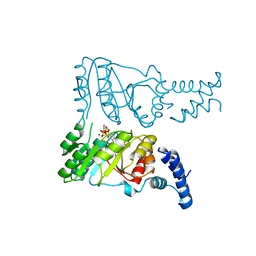 | |
2PX0
 
 | | Crystal structure of FlhF complexed with GMPPNP/Mg(2+) | | 分子名称: | Flagellar biosynthesis protein flhF, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Bange, G, Wild, K, Sinning, I. | | 登録日 | 2007-05-14 | | 公開日 | 2007-09-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4WSN
 
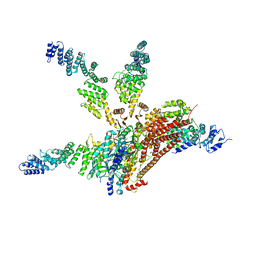 | | Crystal structure of the COP9 signalosome, a P1 crystal form | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | 登録日 | 2014-10-28 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (5.5 Å) | | 主引用文献 | Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome.
Nature, 531, 2016
|
|
5JG6
 
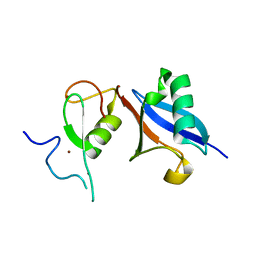 | | APC11-Ubv shows role of noncovalent RING-Ubiquitin interactions in processive multiubiquitination and Ubiquitin chain elongation by APC/C | | 分子名称: | Anaphase-promoting complex subunit 11, Polyubiquitin-B, ZINC ION | | 著者 | Brown, N.G, Zhang, W, Yu, S, Miller, D.J, Sidhu, S.S, Schulman, B.A. | | 登録日 | 2016-04-19 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.0013 Å) | | 主引用文献 | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9U
 
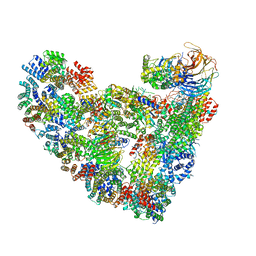 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | 分子名称: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | 著者 | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | 登録日 | 2016-06-11 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9T
 
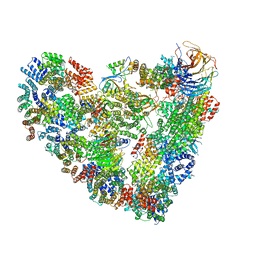 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | 分子名称: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | 著者 | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | 登録日 | 2016-06-11 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
8BU9
 
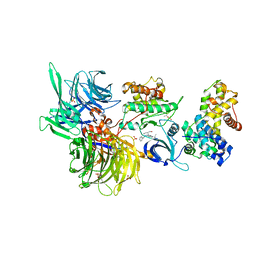 | | Structure of DDB1 bound to roscovitine-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUS
 
 | | Structure of DDB1 bound to DS59-engaged CDK12-cyclin K | | 分子名称: | 1,3-dimethyl-5-[[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)methylamino]purin-2-yl]amino]methyl]pyrazole-4-sulfonamide, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.26 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUG
 
 | | Structure of DDB1 bound to HQ461-engaged CDK12-cyclin K | | 分子名称: | 2-[2-[(6-methylpyridin-2-yl)amino]-1,3-thiazol-4-yl]-~{N}-(5-methyl-1,3-thiazol-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.53 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUH
 
 | | Structure of DDB1 bound to WX3-engaged CDK12-cyclin K | | 分子名称: | 6-[[[2-[[(2~{R})-1-oxidanylbutan-2-yl]amino]-9-propan-2-yl-purin-6-yl]amino]methyl]-3-pyridin-2-yl-1~{H}-pyridin-2-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.79 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUQ
 
 | | Structure of DDB1 bound to DS43-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[[1-(3-chlorophenyl)pyrazol-3-yl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU4
 
 | | Structure of DDB1 bound to DS22-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUC
 
 | | Structure of DDB1 bound to dCeMM3-engaged CDK12-cyclin K | | 分子名称: | 2-(1~{H}-benzimidazol-2-ylsulfanyl)-~{N}-(5-chloranylpyridin-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUT
 
 | | Structure of DDB1 bound to DS61-engaged CDK12-cyclin K | | 分子名称: | 2-[[6-[[4-(2-hydroxyethyloxy)phenyl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU6
 
 | | Structure of DDB1 bound to DS55-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUR
 
 | | Structure of DDB1 bound to DS50-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
