1SM9
 
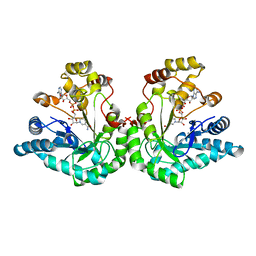 | | Crystal Structure Of An Engineered K274RN276D Double Mutant of Xylose Reductase From Candida Tenuis Optimized To Utilize NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, xylose reductase | | Authors: | Petschacher, B, Leitgeb, S, Kavanagh, K.L, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-03-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography.
Biochem.J., 385, 2005
|
|
1YE4
 
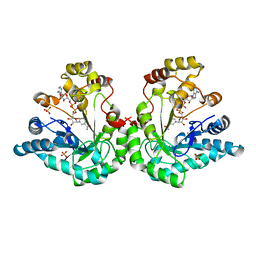 | | Crystal structure of the Lys-274 to Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD+ | | Descriptor: | NAD(P)H-dependent D-xylose reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Leitgeb, S, Petschacher, B, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fine tuning of coenzyme specificity in family 2 aldo-keto reductases revealed by crystal structures of the Lys-274-->Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD(+) and NADP(+).
FEBS Lett., 579, 2005
|
|
1YE6
 
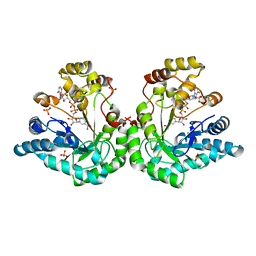 | | Crystal structure of the Lys-274 to Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NADP+ | | Descriptor: | NAD(P)H-dependent D-xylose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Leitgeb, S, Petschacher, B, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fine tuning of coenzyme specificity in family 2 aldo-keto reductases revealed by crystal structures of the Lys-274-->Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD(+) and NADP(+).
Febs Lett., 579, 2005
|
|
2NLX
 
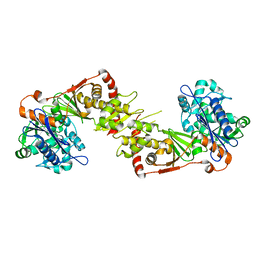 | |
2ITM
 
 | | Crystal structure of the E. coli xylulose kinase complexed with xylulose | | Descriptor: | AMMONIUM ION, D-XYLULOSE, SULFATE ION, ... | | Authors: | di Luccio, E, Voegtli, J, Wilson, D.K. | | Deposit date: | 2006-10-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and kinetic studies of induced fit in xylulose kinase from Escherichia coli.
J.Mol.Biol., 365, 2007
|
|
