3BFD
 
 | |
1W5D
 
 | | Crystal structure of PBP4a from Bacillus subtilis | | 分子名称: | CALCIUM ION, PENICILLIN-BINDING PROTEIN | | 著者 | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | 登録日 | 2004-08-06 | | 公開日 | 2005-12-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Bacillus Subtilis Penicillin-Binding Protein 4A, and its Complex with a Peptidoglycan Mimetic Peptide.
J.Mol.Biol., 371, 2007
|
|
1W8Q
 
 | | Crystal Structure of the DD-Transpeptidase-carboxypeptidase from Actinomadura R39 | | 分子名称: | COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, SULFATE ION | | 著者 | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | 登録日 | 2004-09-24 | | 公開日 | 2005-06-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structure of the Actinomadura R39 Dd-Peptidase Reveals New Domains in Penicillin-Binding Proteins.
J.Biol.Chem., 280, 2005
|
|
1W8Y
 
 | | Crystal structure of the nitrocefin acyl-DD-peptidase from Actinomadura R39. | | 分子名称: | (2R)-2-{(1R)-2-OXO-1-[(2-THIENYLACETYL)AMINO]ETHYL}-5,6-DIHYDRO-2H-1,3-THIAZINE-4-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase, MAGNESIUM ION, ... | | 著者 | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | 登録日 | 2004-10-01 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the Actinomadura R39 Dd- Peptidase Reveals New Domains in Penicillin- Binding Proteins.
J.Biol.Chem., 280, 2005
|
|
3D30
 
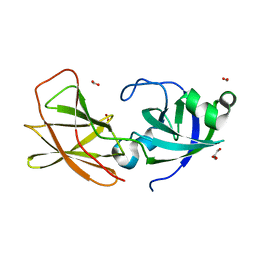 | | Structure of an expansin like protein from Bacillus Subtilis at 1.9A resolution | | 分子名称: | Expansin like protein, FORMIC ACID, GLYCEROL | | 著者 | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Joris, B, Charlier, P. | | 登録日 | 2008-05-09 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3D2Z
 
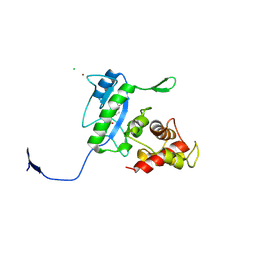 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the product L-Ala-D-gamma-Glu-L-Lys | | 分子名称: | CHLORIDE ION, L-Ala-D-gamma-Glu-L-Lys peptide, N-acetylmuramoyl-L-alanine amidase amiD, ... | | 著者 | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | 登録日 | 2008-05-09 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3D2Y
 
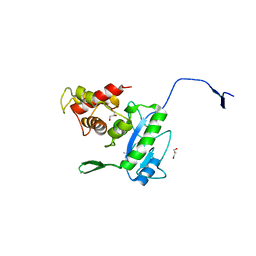 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | 分子名称: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | 著者 | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | 登録日 | 2008-05-09 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
8B7F
 
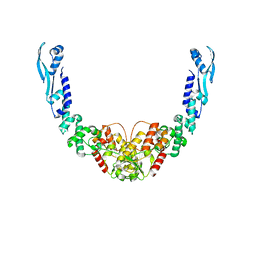 | |
2J9P
 
 | | Crystal structure of the Bacillus subtilis PBP4a, and its complex with a peptidoglycan mimetic peptide. | | 分子名称: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANINE, D-alanyl-D-alanine carboxypeptidase DacC | | 著者 | Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | 登録日 | 2006-11-15 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the Bacillus subtilis penicillin-binding protein 4a, and its complex with a peptidoglycan mimetic peptide.
J. Mol. Biol., 371, 2007
|
|
3IG0
 
 | |
3IFZ
 
 | | crystal structure of the first part of the Mycobacterium tuberculosis DNA gyrase reaction core: the breakage and reunion domain at 2.7 A resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit A, SODIUM ION | | 著者 | Piton, J, Aubry, A, Delarue, M, Mayer, C. | | 登録日 | 2009-07-27 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase.
Plos One, 5, 2010
|
|
