4GPK
 
 | | Crystal structure of NprR in complex with its cognate peptide NprX | | Descriptor: | NprR, NprX peptide | | Authors: | Zouhir, S, Guimaraes, B, Perchat, S, Nicaise, M, Lereclus, D, Nessler, S. | | Deposit date: | 2012-08-21 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR.
Nucleic Acids Res., 41, 2013
|
|
5G31
 
 | |
6S99
 
 | | Dimethylated fusion protein of RSL and trimeric coiled coil in complex with cucurbit[7]uril | | Descriptor: | 4dzn-RSL,Fucose-binding lectin protein,Fucose-binding lectin protein, beta-D-mannopyranose, cucurbit[7]uril | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2019-07-11 | | Release date: | 2020-08-26 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 2021
|
|
6SU0
 
 | | Crystal structure of dimethylated RSLex in complex with cucurbit[7]uril | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, cucurbit[7]uril, ... | | Authors: | Guagnini, F, Engilberge, S, Crowley, P.B. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineered assembly of a protein-cucurbituril biohybrid.
Chem.Commun.(Camb.), 56, 2020
|
|
3V11
 
 | |
3F2M
 
 | | Urate oxidase complexed with 8-azaxanthine at 150 MPa | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Girard, E, Kahn, R, Fourme, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function perturbation and dissociation of tetrameric urate oxidase by high hydrostatic pressure.
Biophys.J., 98, 2010
|
|
3UKG
 
 | | Crystal structure of Rap1/DNA complex | | Descriptor: | CALCIUM ION, DNA-binding protein RAP1, telomeric DNA | | Authors: | Matot, B, Le Bihan, Y.-V, Gasparini, S, LeDu, M.H. | | Deposit date: | 2011-11-09 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The orientation of the C-terminal domain of the Saccharomyces cerevisiae Rap1 protein is determined by its binding to DNA.
Nucleic Acids Res., 40, 2012
|
|
2N0C
 
 | | NMR structure of Neuromedin C in 10% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0B
 
 | | NMR structure of Neuromedin C in aqueous solution | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0F
 
 | | NMR structure of Neuromedin C in 60% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0E
 
 | | NMR structure of Neuromedin C in 40% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0D
 
 | | NMR structure of Neuromedin C in 25% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0H
 
 | | NMR structure of Neuromedin C in presence of SDS micelles | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0G
 
 | | NMR structure of Neuromedin C in 90% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
6STZ
 
 | |
6GD7
 
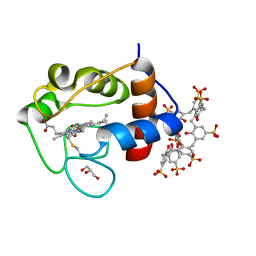 | | Cytochrome c in complex with Sulfonato-calix[8]arene, H3 form with PEG | | Descriptor: | Cytochrome c iso-1, GLYCEROL, HEME C, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GD8
 
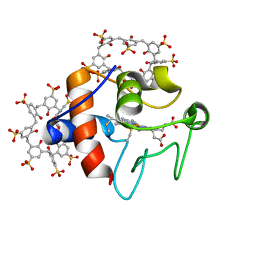 | | Cytochrome c in complex with Sulfonato-calix[8]arene, P31 form | | Descriptor: | Cytochrome c iso-1, HEME C, sulfonato-calix[8]arene | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GD9
 
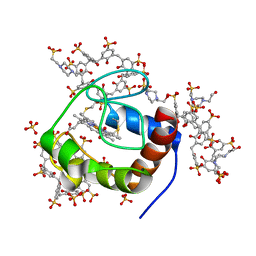 | | Cytochrome c in complex with Sulfonato-calix[8]arene, P43212 form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytochrome c iso-1, HEME C, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GD6
 
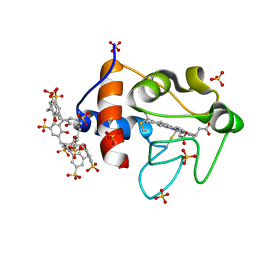 | | Cytochrome c in complex with Sulfonato-calix[8]arene, H3 form with ammonium sulfate | | Descriptor: | Cytochrome c iso-1, HEME C, SULFATE ION, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8RQP
 
 | | In meso structure of the alginate exporter, AlgE, from Pseudomonas aeruginosa in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), Alginate production protein AlgE, ... | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQR
 
 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (S-form), Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQQ
 
 | | In meso structure of the adenosine A2a G protein-coupled receptor, A2aR, in 7.10 monoacylglycerol | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), ... | | Authors: | Smithers, L, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
4DO2
 
 | |
7P2H
 
 | | Dimethylated fusion protein of RSL and mussel adhesion peptide (Mefp) in complex with cucurbit[7]uril, H3 sheet assembly | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SODIUM ION, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 27, 2021
|
|
7P2I
 
 | | Dimethylated fusion protein of RSL and nucleoporin peptide (Nup) in complex with cucurbit[7]uril, F432 cage assembly | | Descriptor: | Fucose-binding lectin protein, cucurbit[7]uril, methyl alpha-L-fucopyranoside | | Authors: | Guagnini, F, Ramberg, K, Crowley, P.B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 27, 2021
|
|
