2FCD
 
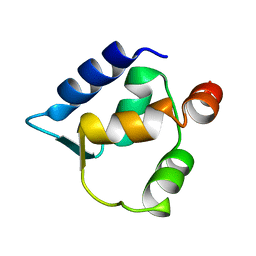 | | Solution structure of N-lobe Myosin Light Chain from Saccharomices cerevisiae | | Descriptor: | Myosin light chain 1 | | Authors: | Cicero, D.O, Pennestri, M, Contessa, G.M, Paci, M, Ragnini-Wilson, A, Melino, S. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of the myosin light chain Mlc1p with the myosin V Myo2p IQ motifs.
J.Biol.Chem., 282, 2007
|
|
2FCE
 
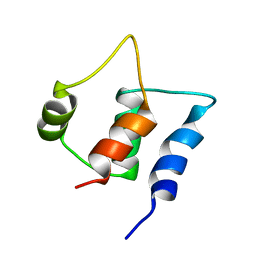 | | Solution structure of C-lobe Myosin Light Chain from Saccharomices cerevisiae | | Descriptor: | Myosin light chain 1 | | Authors: | Cicero, D.O, Pennestri, M, Contessa, G.M, Paci, M, Ragnini-Wilson, A, Melino, S. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of the myosin light chain Mlc1p with the myosin V Myo2p IQ motifs.
J.Biol.Chem., 282, 2007
|
|
2K1Q
 
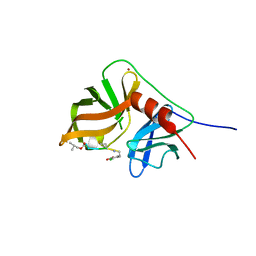 | | NMR structure of hepatitis c virus ns3 serine protease complexed with the non-covalently bound phenethylamide inhibitor | | Descriptor: | NS3 PROTEASE, PHENETHYLAMIDE, ZINC ION | | Authors: | Eliseo, T, Gallo, M, Pennestri, M, Bazzo, R, Cicero, D.O. | | Deposit date: | 2008-03-13 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Binding of a noncovalent inhibitor exploiting the S' region stabilizes the hepatitis C virus NS3 protease conformation in the absence of cofactor.
J.Mol.Biol., 385, 2009
|
|
