8UR9
 
 | | Crystal Structure of the SARS-CoV-2 Main Protease in Complex with Compound 61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Papini, C, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-10-25 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proof-of-concept studies with a computationally designed M pro inhibitor as a synergistic combination regimen alternative to Paxlovid.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BO8
 
 | | Crystal Structure of T21I SARS-CoV-2 Main Protease in Complex with GC376 | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Papini, C, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BO1
 
 | | Crystal Structure of T190I SARS-CoV-2 Main Protease in Complex with Compound Mpro61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Papini, C, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BO6
 
 | | Crystal Structure of A173V/L50F SARS-CoV-2 Main Protease in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Papini, C, Kenneson, J, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BOE
 
 | |
9BOC
 
 | |
9BNV
 
 | |
9BNY
 
 | |
9BNU
 
 | | Crystal Structure of T190I SARS-CoV-2 Main Protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Papini, C, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BO0
 
 | | Crystal Structure of T21I SARS-CoV-2 Main Protease in Complex with Compound Mpro61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase | | Authors: | Papini, C, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BO9
 
 | | Crystal Structure of T190I SARS-CoV-2 Main Protease in Complex with GC376 | | Descriptor: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Papini, C, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BO7
 
 | | Crystal Structure of L50F SARS-CoV-2 Main Protease in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Papini, C, Kenneson, J, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BNX
 
 | |
9BO2
 
 | | Crystal Structure of A173V/L50F SARS-CoV-2 Main Protease in Complex with Compound Mpro61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Papini, C, Kenneson, J, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BNT
 
 | | Crystal Structure of T21I SARS-CoV-2 Main Protease | | Descriptor: | 3C-like proteinase | | Authors: | Papini, C, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BO4
 
 | | Crystal Structure of T21I SARS-CoV-2 Main Protease in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Papini, C, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BNW
 
 | |
9BO5
 
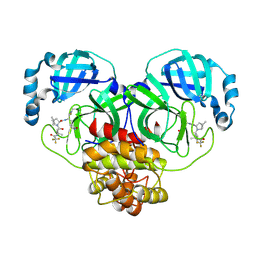 | | Crystal Structure of T190I SARS-CoV-2 Main Protease in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Papini, C, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BOA
 
 | |
9BOB
 
 | |
9BOD
 
 | |
9BO3
 
 | | Crystal Structure of E166V SARS-CoV-2 Main Protease in Complex with Compound Mpro61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Papini, C, Kenneson, J, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
6YWW
 
 | | MeCP2 is a microsatellite binding protein that protects CA repeats from nucleosome invasion | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*AP*AP*TP*TP*GP*TP*GP*TP*GP*TP*GP*TP*GP*CP*AP*G)-3'), DNA/RNA (5'-D(*TP*CP*TP*GP*CP*AP*CP*A)-R(P*(5HC))-D(P*AP*CP*AP*CP*AP*AP*TP*TP*AP*TP*A)-3'), Truncated methyl CpG binding protein 2 transcript 1 | | Authors: | Ibrahim, A, Papin, C, Mohideen-Abdul, K, Gras, S.L, Stoll, I, Bronner, C, Dimitrov, S, Klaholz, B.P, Hamiche, A. | | Deposit date: | 2020-04-30 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | MeCP2 is a microsatellite binding protein that protects CA repeats from nucleosome invasion.
Science, 372, 2021
|
|
5FUG
 
 | | Crystal structure of a human YL1-H2A.Z-H2B complex | | Descriptor: | HISTONE H2A.Z, HISTONE H2B TYPE 1-J, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 72 HOMOLOG | | Authors: | Latrick, C.M, Marek, M, Ouararhni, K, Papin, C, Stoll, I, Ignatyeva, M, Obri, A, Ennifar, E, Dimitrov, S, Romier, C, Hamiche, A. | | Deposit date: | 2016-01-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis and Specificity of H2A.Z-H2B Recognition and Deposition by the Histone Chaperone Yl1
Nat.Struct.Mol.Biol., 23, 2016
|
|
4CAY
 
 | | Crystal structure of a human Anp32e-H2A.Z-H2B complex | | Descriptor: | ACIDIC LEUCINE-RICH NUCLEAR PHOSPHOPROTEIN 32 FAMILY MEMBER E, HISTONE H2A.Z, HISTONE H2B TYPE 1-J | | Authors: | Obri, A, Ouararhni, K, Papin, C, Diebold, M.-L, Padmanabhan, K, Marek, M, Stoll, I, Roy, L, Reilly, P.T, Mak, T.W, Dimitrov, S, Romier, C, Hamiche, A. | | Deposit date: | 2013-10-09 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Anp32E is a Histone Chaperone that Removes H2A.Z from Chromatin
Nature, 648, 2014
|
|
