2JK0
 
 | | Structural and functional insights into Erwinia carotovora L- asparaginase | | Descriptor: | ASPARTIC ACID, L-ASPARAGINASE | | Authors: | Papageorgiou, A.C, Posypanova, G.A, Andersson, C.S, Sokolov, N.N, Krasotkina, J. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Insights Into Erwinia Carotovora L-Asparaginase.
FEBS J., 275, 2008
|
|
5M60
 
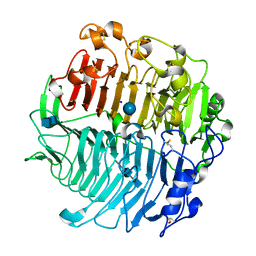 | | Chaetomium thermophilum beta-1-3-glucanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, SODIUM ION, ... | | Authors: | Papageorgiou, A.C, Chen, J, Li, D. | | Deposit date: | 2016-10-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and biological implications of a glycoside hydrolase family 55 beta-1,3-glucanase from Chaetomium thermophilum.
Biochim. Biophys. Acta, 1865, 2017
|
|
1UNS
 
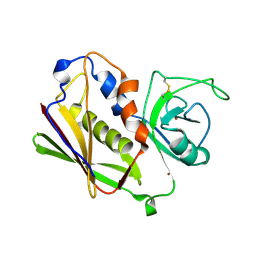 | | IDENTIFICATION OF A SECONDARY ZINC-BINDING SITE IN STAPHYLOCOCCAL ENTEROTOXIN C2: IMPLICATIONS FOR SUPERANTIGEN RECOGNITION | | Descriptor: | ENTEROTOXIN TYPE C-2, ZINC ION | | Authors: | Papageorgiou, A.C, Baker, M.D, McLeod, J.D, Goda, S, Sansom, D.M, Tranter, H.S, Acharya, K.R. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Secondary Zinc-Binding Site in Staphylococcal Enterotoxin C2: Implications for Superantigen Recognition
J.Biol.Chem., 279, 2004
|
|
2VTV
 
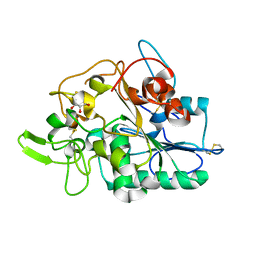 | | PhaZ7 depolymerase from Paucimonas lemoignei | | Descriptor: | GLYCEROL, PHB depolymerase PhaZ7 | | Authors: | Papageorgiou, A.C, Hermawan, S, Singh, C.B, Jendrossek, D. | | Deposit date: | 2008-05-16 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of poly(3-hydroxybutyrate) hydrolysis by PhaZ7 depolymerase from Paucimonas lemoignei.
J. Mol. Biol., 382, 2008
|
|
3SEB
 
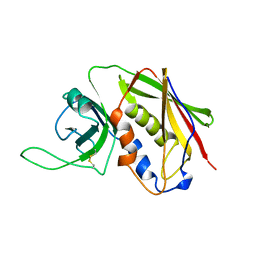 | | STAPHYLOCOCCAL ENTEROTOXIN B | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN B | | Authors: | Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1997-11-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of microbial superantigen staphylococcal enterotoxin B at 1.5 A resolution: implications for superantigen recognition by MHC class II molecules and T-cell receptors.
J.Mol.Biol., 277, 1998
|
|
1A4Y
 
 | | RIBONUCLEASE INHIBITOR-ANGIOGENIN COMPLEX | | Descriptor: | ANGIOGENIN, RIBONUCLEASE INHIBITOR | | Authors: | Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1998-02-08 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular recognition of human angiogenin by placental ribonuclease inhibitor--an X-ray crystallographic study at 2.0 A resolution.
EMBO J., 16, 1997
|
|
1B1Z
 
 | | STREPTOCOCCAL PYROGENIC EXOTOXIN A1 | | Descriptor: | PROTEIN (TOXIN) | | Authors: | Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1998-11-24 | | Release date: | 1999-11-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for the recognition of superantigen streptococcal pyrogenic exotoxin A (SpeA1) by MHC class II molecules and T-cell receptors.
EMBO J., 18, 1999
|
|
5M5Z
 
 | | Chaetomium thermophilum beta-1-3-glucanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, ... | | Authors: | Papageorgiou, A.C, Chen, J, Li, D. | | Deposit date: | 2016-10-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure and biological implications of a glycoside hydrolase family 55 beta-1,3-glucanase from Chaetomium thermophilum.
Biochim. Biophys. Acta, 1865, 2017
|
|
1DUE
 
 | |
1DT2
 
 | |
1DUA
 
 | |
6ZS1
 
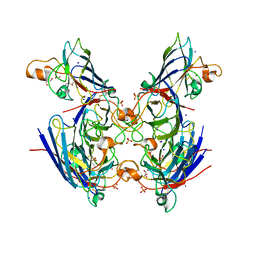 | | Chaetomium thermophilum CuZn-superoxide dismutase | | Descriptor: | COPPER (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Papageorgiou, A.C, Mohsin, I. | | Deposit date: | 2020-07-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of a Cu,Zn Superoxide Dismutase From the Thermophilic Fungus Chaetomium thermophilum.
Protein Pept.Lett., 28, 2021
|
|
1QIL
 
 | |
7BEU
 
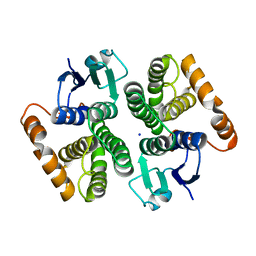 | | Human glutathione transferase M1-1 | | Descriptor: | Glutathione S-transferase Mu 1, SODIUM ION | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2020-12-28 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Ligandability Assessment of Human Glutathione Transferase M1-1 Using Pesticides as Chemical Probes.
Int J Mol Sci, 23, 2022
|
|
7ZA4
 
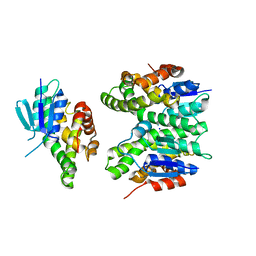 | | GSTF sh155 mutant | | Descriptor: | Glutathione transferase, SODIUM ION | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Directed Evolution of Phi Class Glutathione Transferases Involved in Multiple-Herbicide Resistance of Grass Weeds and Crops.
Int J Mol Sci, 23, 2022
|
|
7ZA5
 
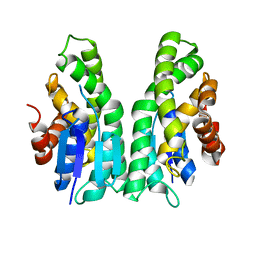 | | GSTF sh101 mutant | | Descriptor: | Glutathione transferase | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Directed Evolution of Phi Class Glutathione Transferases Involved in Multiple-Herbicide Resistance of Grass Weeds and Crops.
Int J Mol Sci, 23, 2022
|
|
8R37
 
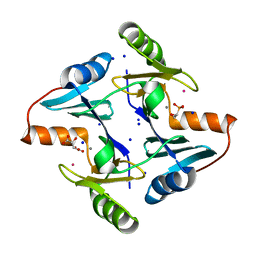 | | Klebsiella pneumoniae fosfomycin-resistance protein (FosAKP) | | Descriptor: | FOSFOMYCIN, FosA family fosfomycin resistance glutathione transferase, L(+)-TARTARIC ACID, ... | | Authors: | Papageorgiou, A.C, Varotsou, C, Labrou, N.E. | | Deposit date: | 2023-11-08 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Studies of Klebsiella pneumoniae Fosfomycin-Resistance Protein and Its Application for the Development of an Optical Biosensor for Fosfomycin Determination.
Int J Mol Sci, 25, 2023
|
|
6YRO
 
 | | Streptococcus suis SadP mutant - N285D | | Descriptor: | GLYCEROL, SODIUM ION, SadP | | Authors: | Papageorgiou, A.C, Haataja, S. | | Deposit date: | 2020-04-20 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The binding mechanism of the virulence factor Streptococcus suis adhesin P subtype to globotetraosylceramide is associated with systemic disease.
J.Biol.Chem., 295, 2020
|
|
6GHF
 
 | | Crystal structure of a GST variant | | Descriptor: | PvGmGSTUG | | Authors: | Papageorgiou, A.C, Chronopoulou, E.G, Labrou, N.E. | | Deposit date: | 2018-05-07 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Expanding the Plant GSTome Through Directed Evolution: DNA Shuffling for the Generation of New Synthetic Enzymes With Engineered Catalytic and Binding Properties.
Front Plant Sci, 9, 2018
|
|
2QIL
 
 | |
8BHC
 
 | | K141H and S142H double mutant of hGSTA1-1 | | Descriptor: | Glutathione S-transferase A1 | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2022-10-31 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A Key Role in Catalysis and Enzyme Thermostability of a Conserved Helix H5 Motif of Human Glutathione Transferase A1-1.
Int J Mol Sci, 24, 2023
|
|
8C5D
 
 | | Glutathione transferase P1-1 from Mus musculus | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2023-01-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Inhibition Analysis and High-Resolution Crystal Structure of Mus musculus Glutathione Transferase P1-1.
Biomolecules, 13, 2023
|
|
8C4D
 
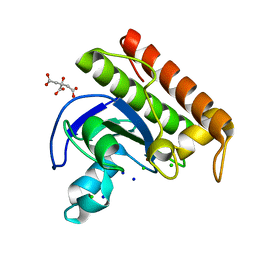 | | N-acetylmuramoyl-L-alanine amidase from Enterococcus faecium prophage genome | | Descriptor: | CHLORIDE ION, CITRATE ANION, N-acetylmuramoyl-L-alanine amidase, ... | | Authors: | Papageorgiou, A.C, Premetis, G.E, Labrou, N.E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional features of a broad-spectrum prophage-encoded enzybiotic from Enterococcus faecium.
Sci Rep, 13, 2023
|
|
8BHE
 
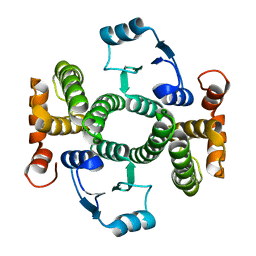 | | K141H and S142H double mutant of hGSTA1-1 | | Descriptor: | Glutathione S-transferase A1 | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2022-10-31 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A Key Role in Catalysis and Enzyme Thermostability of a Conserved Helix H5 Motif of Human Glutathione Transferase A1-1.
Int J Mol Sci, 24, 2023
|
|
1STE
 
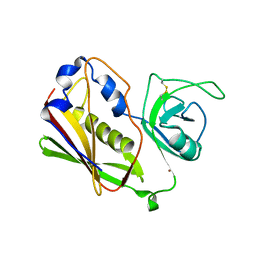 | |
