4P7V
 
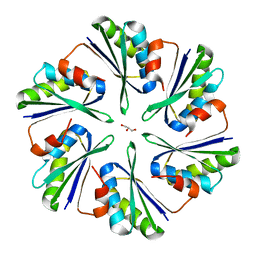 | | Structural insights into higher-order assembly and function of the bacterial microcompartment protein PduA | | Descriptor: | GLYCEROL, Polyhedral bodies | | Authors: | Pang, A, Frank, S, Brown, I.R, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into Higher Order Assembly and Function of the Bacterial Microcompartment Protein PduA.
J.Biol.Chem., 289, 2014
|
|
4P7T
 
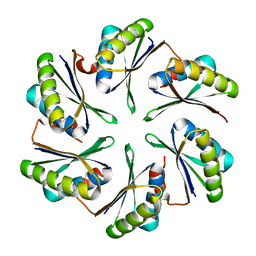 | |
3PAC
 
 | |
4FAY
 
 | | Crystal structure of a trimeric bacterial microcompartment shell protein PduB with glycerol metabolites | | Descriptor: | ACETATE ION, GLYCEROL, Microcompartments protein | | Authors: | Pang, A.H, Prentice, M.B, Pickersgill, R.W. | | Deposit date: | 2012-05-22 | | Release date: | 2012-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4QIF
 
 | | Crystal Structure of PduA with edge mutation K26A and pore mutation S40H | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Pang, A.H, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2014-05-30 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9951 Å) | | Cite: | Selective molecular transport through the protein shell of a bacterial microcompartment organelle.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
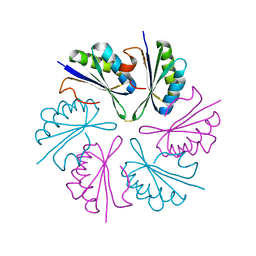
4QIG
 
 | |
4RBT
 
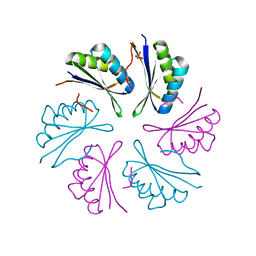 | |
4RBV
 
 | |
4RBU
 
 | |
8UHO
 
 | | Crystal structure of SARS CoV-2 3CL protease in complex with GSK4365096A | | Descriptor: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-[(2S,3Z)-1-[(2S)-oxolan-2-yl]-3-(2-oxooxolan-3-ylidene)propan-2-yl]-L-leucinamide | | Authors: | Concha, N.O, Williams, S.P. | | Deposit date: | 2023-10-09 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
8ULD
 
 | | SARA CoV-2 3C-like protease in complex with GSK3487016A | | Descriptor: | 1,2-ETHANEDIOL, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-{(2S,3R)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]-4-[(propan-2-yl)amino]butan-2-yl}-L-leucinamide, Replicase polyprotein 1a | | Authors: | Williams, S.P, Concha, N.O. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
8UIF
 
 | | Crystal structure of SARS CoV-2 3CL protease in complex with GSK4365096A | | Descriptor: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-{(2R)-1-[(2S)-oxolan-2-yl]-3-[(3S)-2-oxooxolan-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Concha, N.O, Williams, S.P. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
8UIA
 
 | |
6ODL
 
 | | Crystal structure of GluN2A agonist binding domain with 4-butyl-(S)-CCG-IV | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]-1-butylcyclopropane-1-carboxylic acid, Glutamate receptor ionotropic, NMDA 2A,Glutamate receptor ionotropic, ... | | Authors: | Mou, T.C, Clausen, R.P, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stereoselective synthesis of novel 2'-(S)-CCG-IV analogues as potent NMDA receptor agonists.
Eur.J.Med.Chem., 212, 2021
|
|
