6QHW
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 4512 ms | | Descriptor: | Fluoroacetate dehalogenase, GLYCOLIC ACID, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QHS
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 564 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
1X1Z
 
 | | Orotidine 5'-monophosphate decarboxylase (odcase) complexed with BMP (produced from 6-cyanoump) | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fujihashi, M, Bello, A.M, Poduch, E, Wei, L, Annedi, S.C, Pai, E.F, Kotra, L.P. | | Deposit date: | 2005-04-15 | | Release date: | 2005-12-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An unprecedented twist to ODCase catalytic activity
J.Am.Chem.Soc., 127, 2005
|
|
6QI0
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 9024 ms | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QHV
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 100 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QKS
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Apo | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
2EAW
 
 | |
2E3T
 
 | | Crystal structure of rat xanthine oxidoreductase mutant (W335A and F336L) | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Asai, R, Nishino, T, Matsumura, T, Okamoto, K, Pai, E.F, Nishino, T. | | Deposit date: | 2006-11-28 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Two mutations convert mammalian xanthine oxidoreductase to highly superoxide-productive xanthine oxidase
J.Biochem.(Tokyo), 141, 2007
|
|
2E6Y
 
 | | Covalent complex of orotidine 5'-monophosphate decarboxylase (ODCase) with 6-Iodo-UMP | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fujihashi, M, Bello, A.M, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Potent, Covalent Inhibitor of Orotidine 5'-Monophosphate Decarboxylase with Antimalarial Activity.
J.Med.Chem., 50, 2007
|
|
1YNB
 
 | | crystal structure of genomics APC5600 | | Descriptor: | hypothetical protein AF1432 | | Authors: | Dong, A, Skarina, T, Savchenko, A, Pai, E.F, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of genomics AF1432 by Sulfur SAD methods
To be Published
|
|
1YQE
 
 | | Crystal Structure of Conserved Protein of Unknown Function AF0625 | | Descriptor: | Hypothetical UPF0204 protein AF0625, PYROPHOSPHATE 2- | | Authors: | Liu, Y, Skarina, T, Dong, A, Kudritskam, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-01 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein AF0625
To be Published
|
|
1YXO
 
 | | Crystal Structure of pyridoxal phosphate biosynthetic protein PdxA PA0593 | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, ETHANOL, MAGNESIUM ION | | Authors: | Liu, Y, Xu, X, Dong, A, Kudritskam, M, Savchenko, A, Pai, E.F, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-05 | | Last modified: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of pyridoxal phosphate biosynthetic protein PdxA PA0593
To be Published
|
|
2ESH
 
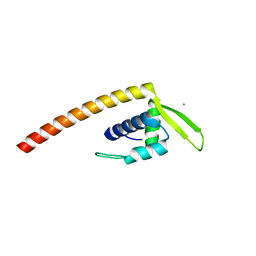 | | Crystal Structure of Conserved Protein of Unknown Function TM0937- a Potential Transcriptional Factor | | Descriptor: | CALCIUM ION, conserved hypothetical protein TM0937 | | Authors: | Liu, Y, Bochkareva, E, Zheng, H, Xu, X, Nocek, B, Lunin, V, Edward, A, Pai, E.F, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein TM0937
To be Published
|
|
2F5A
 
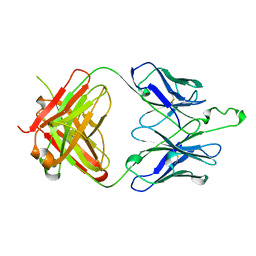 | | CRYSTAL STRUCTURE OF FAB' FROM THE HIV-1 NEUTRALIZING ANTIBODY 2F5 | | Descriptor: | PROTEIN (ANTIBODY 2F5 (HEAVY CHAIN)), PROTEIN (ANTIBODY 2F5 (LIGHT CHAIN)) | | Authors: | Bryson, S, Julien, J.P, Hynes, R.C, Pai, E.F. | | Deposit date: | 1999-04-08 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications.
J.Virol., 83, 2009
|
|
2F5B
 
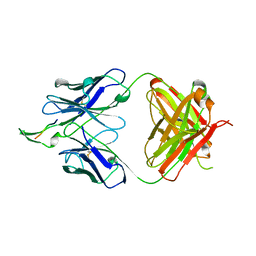 | | CRYSTAL STRUCTURE OF FAB' FROM THE HIV-1 NEUTRALIZING ANTIBODY 2F5 IN COMPLEX WITH ITS GP41 EPITOPE | | Descriptor: | PROTEIN (ANTIBODY 2F5 (HEAVY CHAIN)), PROTEIN (ANTIBODY 2F5 (LIGHT CHAIN)), PROTEIN (GP41 EPITOPE) | | Authors: | Bryson, S, Julien, J.P, Hynes, R.C, Pai, E.F. | | Deposit date: | 1999-04-09 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications.
J.Virol., 83, 2009
|
|
1ZK7
 
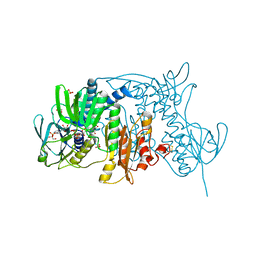 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase, ... | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-05-02 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
2FV2
 
 | |
2FTP
 
 | | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SODIUM ION, hydroxymethylglutaryl-CoA lyase | | Authors: | Xiao, T, Evdokimova, E, Liu, Y, Kudritska, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-24 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa
To be Published
|
|
1ZX9
 
 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mercuric reductase | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-06-07 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
2GUU
 
 | | crystal structure of Plasmodium vivax orotidine 5-monophosphate decarboxylase with 6-aza-UMP bound | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, ODcase | | Authors: | Dong, A, Lew, J, Zhao, Y, Sundararajan, E, Wasney, G, Vedadi, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Pai, E.F, Kotra, L, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of Plasmodium vivax orotidine 5-monophosphate decarboxylase with 6-aza-UMP bound
To be Published
|
|
2AZP
 
 | | Crystal Structure of PA1268 Solved by Sulfur SAD | | Descriptor: | hypothetical protein PA1268 | | Authors: | Liu, Y, Gorodichtchenskaia, E, Skarina, T, Yang, C, Joachimiak, A, Edwards, A, Pai, E.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of PA1268 Solved by Sulfur SAD
To be Published
|
|
2HN2
 
 | |
2ZZ7
 
 | |
2ZZ5
 
 | |
2ZZ3
 
 | |
