3X28
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-12 | | Release date: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Reaction intermediate of nitrile hydratase determined by time-resolved crystallography reveals the cysteine-sulfenic acid ligand to be a catalytic nucleophile
To be Published
|
|
3A08
 
 | | Structure of (PPG)4-OOG-(PPG)4, monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Wu, G, Noguchi, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-03-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Two crystal modifications of (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 reveal the puckering preference of Hyp(X) in the Hyp(X):Hyp(Y) and Hyp(X):Pro(Y) stacking pairs in collagen helices.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3ABN
 
 | | Crystal structure of (Pro-Pro-Gly)4-Hyp-Asp-Gly-(Pro-Pro-Gly)4 at 1.02 A | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Shimura, M, Kawaguchi, T, Noguchi, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-12-16 | | Release date: | 2010-12-01 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the collagen model peptide (Pro-Pro-Gly)4 -Hyp-Asp-Gly-(Pro-Pro-Gly)4 at 1.0 angstrom resolution.
Biopolymers, 99, 2013
|
|
3A14
 
 | | Crystal structure of DXR from Thermotoga maritima, in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MAGNESIUM ION, ... | | Authors: | Takenoya, M, Ohtaki, A, Noguchi, K, Sasaki, Y, Ohsawa, K, Yohda, M, Yajima, S. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase from the hyperthermophile Thermotoga maritima for insights into the coordination of conformational changes and an inhibitor binding.
J.Struct.Biol., 2010
|
|
3A1K
 
 | | Crystal structure of Rhodococcus sp. N771 Amidase | | Descriptor: | Amidase | | Authors: | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | Deposit date: | 2009-04-09 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and characterization of amidase from Rhodococcus sp. N-771: Insight into the molecular mechanism of substrate recognition
Biochim.Biophys.Acta, 1804, 2010
|
|
3A06
 
 | | Crystal structure of DXR from Thermooga maritia, in complex with fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, ... | | Authors: | Takenoya, M, Ohtaki, A, Noguchi, K, Sasaki, Y, Ohsawa, K, Yohda, M, Yajima, S. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase from the hyperthermophile Thermotoga maritima for insights into the coordination of conformational changes and an inhibitor binding
J.Struct.Biol., 170, 2010
|
|
3A1I
 
 | | Crystal structure of Rhodococcus sp. N-771 Amidase complexed with Benzamide | | Descriptor: | Amidase, BENZAMIDE | | Authors: | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | Deposit date: | 2009-04-03 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Characterization of Amidase from Rhodococcus sp. N-771: Insight into the Molecular Mechanism of Substrate Recognition
Biochim.Biophys.Acta, 2009
|
|
3A8G
 
 | | Crystal structure of Nitrile Hydratase mutant S113A complexed with Trimethylacetonitrile | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
3A8M
 
 | | Crystal structure of Nitrile Hydratase mutant Y72F complexed with Trimethylacetonitrile | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
3A8L
 
 | | Crystal structure of photo-activation state of Nitrile Hydratase mutant S113A | | Descriptor: | FE (III) ION, Nitrile hydratase subunit alpha, Nitrile hydratase subunit beta | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
3A8O
 
 | | Crystal structure of Nitrile Hydratase complexed with Trimethylacetamide | | Descriptor: | 2,2-dimethylpropanamide, FE (III) ION, Nitrile hydratase subunit alpha, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
3A8H
 
 | | Crystal structure of Nitrile Hydratase mutant S113A complexed with Trimethylacetamide | | Descriptor: | 2,2-dimethylpropanamide, FE (III) ION, Nitrile hydratase subunit alpha, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
3VQJ
 
 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, ZINC ION | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-03-24 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3VRK
 
 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase / Thiocyanate complex | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-04-11 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3VYH
 
 | | Crystal structure of aW116R mutant of nitrile hydratase from Pseudonocardia thermophilla | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Yamanaka, Y, Sato, M, Arakawa, T, Namima, S, Hori, S, Ohtaki, A, Noguchi, K, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2012-09-25 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
To be published
|
|
3VYG
 
 | | Crystal structure of Thiocyanate hydrolase mutant R136W | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Yamanaka, Y, Sato, M, Arakawa, T, Namima, S, Hori, S, Ohtaki, A, Noguchi, K, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2012-09-25 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
To be published
|
|
3WE1
 
 | | Crystal structure of Dengue 4 Envelope protein domain III (ED3) | | Descriptor: | Envelope protein E | | Authors: | Elahi, M, Islam, M.M, Noguchi, K, Yohda, M, Toh, H, Kuroda, Y. | | Deposit date: | 2013-06-27 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Computational prediction and experimental characterization of a "size switch type repacking" during the evolution of dengue envelope protein domain III (ED3).
Biochim.Biophys.Acta, 1844, 2014
|
|
2ZJX
 
 | | Bovine pancreatic trypsin inhibitor (BPTI) containing only the [5,55] disulfide bond | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Sohya, S, Noguchi, K, Yohda, M, Kuroda, Y. | | Deposit date: | 2008-03-11 | | Release date: | 2008-10-21 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal structure of an extensively simplified variant of bovine pancreatic trypsin inhibitor in which over one-third of the residues are alanines
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZVX
 
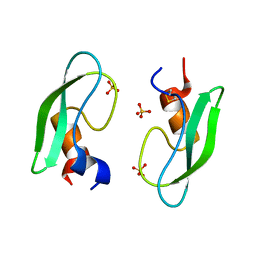 | | Structure of a BPTI-[5,55] variant containing Gly/Val at the 14/38th positions | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Sohya, S, Noguchi, K, Yohda, M, Kuroda, Y. | | Deposit date: | 2008-11-24 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Thermodynamic and structural analysis of highly stabilized BPTIs by single and double mutations
Proteins, 77, 2009
|
|
3AEI
 
 | | Crystal structure of the prefoldin beta2 subunit from Thermococcus strain KS-1 | | Descriptor: | CHLORIDE ION, Prefoldin beta subunit 2, SULFATE ION | | Authors: | Ohtaki, A, Sugano, Y, Sato, T, Noguchi, K, Miyatake, H, Yohda, M. | | Deposit date: | 2010-02-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic Characterization of the Interaction between Prefoldin and Group II Chaperonin
J.Mol.Biol., 399, 2010
|
|
3AUD
 
 | | Simplified BPTI variant with poly Asn amino acid tag (C5N) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUG
 
 | | A simplified BPTI variant with poly Pro amino acid tag (C5P) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Effect of amino acid mutations of protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUB
 
 | | A simplified BPTI variant stabilized by the A14G and A38V substitutions | | Descriptor: | Bovine Pancreatic trypsin inhibitor | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Effect of amino acid mutations of protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUI
 
 | | A simplified BPTI variant with poly Glu amino acid tag (C3E) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUH
 
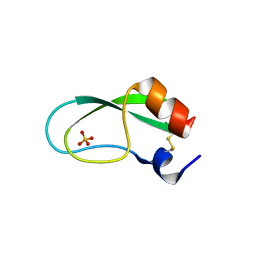 | | A simplified BPTI variant with poly Arg amino acid tag (C3R) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
