3VOT
 
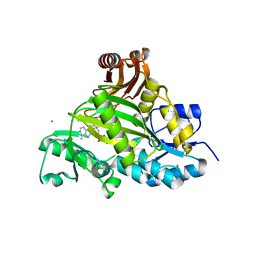 | | Crystal structure of L-amino acid ligase from Bacillus licheniformis | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Suzuki, M, Takahashi, Y, Noguchi, A, Arai, T, Yagasaki, M, Kino, K, Saito, J. | | 登録日 | 2012-02-08 | | 公開日 | 2012-11-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of L-amino-acid ligase from Bacillus licheniformis
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8K5G
 
 | |
8K5H
 
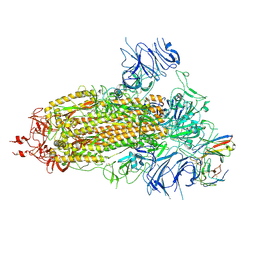 | | Structure of the SARS-CoV-2 BA.1 spike with UT28-RD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | 登録日 | 2023-07-21 | | 公開日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | In silico-designed UT28K with two amino acid substitutions is capable of neutralization of resistant SARS-CoV2 Omicrons BA.1 valiant.
To Be Published
|
|
