5B2G
 
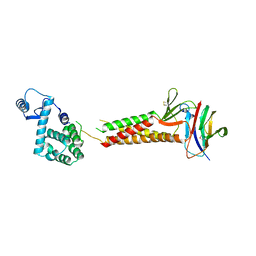 | | Crystal structure of human claudin-4 in complex with C-terminal fragment of Clostridium perfringens enterotoxin | | Descriptor: | Endolysin,Claudin-4, Heat-labile enterotoxin B chain | | Authors: | Shinoda, T, Kimura-Someya, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin
Sci Rep, 6, 2016
|
|
2ZXE
 
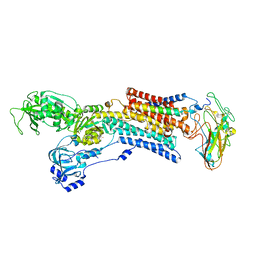 | | Crystal structure of the sodium - potassium pump in the E2.2K+.Pi state | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Shinoda, T, Ogawa, H, Cornelius, F, Toyoshima, C. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the sodium - potassium pump at 2.4 A resolution
Nature, 459, 2009
|
|
6EHM
 
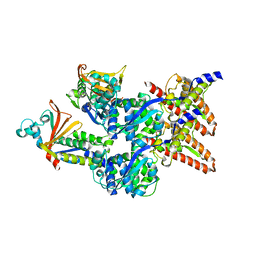 | | Model of the Ebola virus nucleocapsid subunit from recombinant virus-like particles | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
6EHL
 
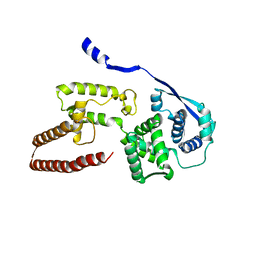 | | Model of the Ebola virus nucleoprotein in recombinant nucleocapsid-like assemblies | | Descriptor: | Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
7YPW
 
 | | Lloviu cuevavirus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Hu, S.F, Fujita-Fujiharu, Y, Sugita, Y, Wendt, L, Muramoto, Y, Nakano, M, Hoenen, T, Noda, T. | | Deposit date: | 2022-08-04 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.0356 Å) | | Cite: | Cryoelectron microscopic structure of the nucleoprotein-RNA complex of the European filovirus, Lloviu virus.
Pnas Nexus, 2, 2023
|
|
7YR8
 
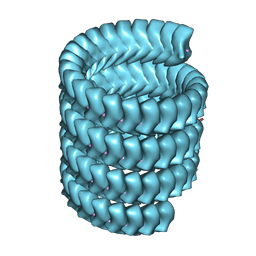 | | Lloviu cuevavirus nucleoprotein(1-450 residues)-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Hu, S.F, Fujita-Fujiharu, Y, Sugita, Y, Wendt, L, Muramoto, Y, Nakano, M, Hoenen, T, Noda, T. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryoelectron microscopic structure of the nucleoprotein-RNA complex of the European filovirus, Lloviu virus.
Pnas Nexus, 2, 2023
|
|
8Y9J
 
 | | Structure of the Ebola virus nucleocapsid subunit | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein, RNA (12-MER) | | Authors: | Fujita-Fujiharu, Y, Hu, S, Hirabayashi, A, Takamatsu, Y, Ng, Y, Houri, K, Muramoto, Y, Nakano, M, Sugita, Y, Noda, T. | | Deposit date: | 2024-02-06 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for Ebola virus nucleocapsid assembly and function regulated by VP24.
Nat Commun, 16, 2025
|
|
7F1M
 
 | | Marburg virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Fujita, F.Y, Sugita, Y, Takamatsu, Y, Houri, K, Muramoto, Y, Nakano, M, Tsunoda, Y, Igarashi, M, Becker, S, Noda, T. | | Deposit date: | 2021-06-09 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Marburg virus nucleoprotein-RNA complex formation.
Nat Commun, 13, 2022
|
|
9JSK
 
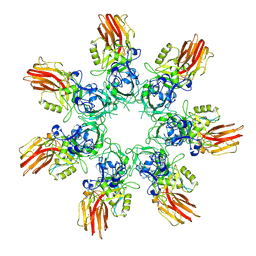 | | Clostridium perfringens iota toxin pore Ib in prepore IV state | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yamada, T, Sugita, Y, Yoshida, T, Noda, T, Tsuge, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
To Be Published
|
|
9JSF
 
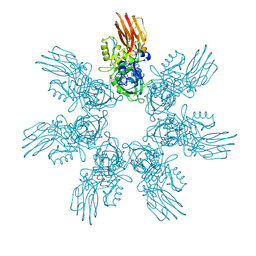 | | Clostridium perfringens iota toxin pore Ib in prepore I state | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yamada, T, Sugita, Y, Yoshida, T, Noda, T, Tsuge, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
To Be Published
|
|
9JSG
 
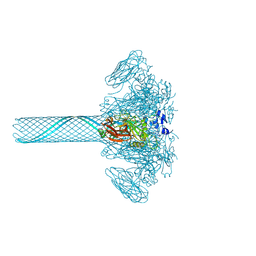 | | Clostridium perfringens iota toxin pore Ib in pore state | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yamada, T, Sugita, Y, Yoshida, T, Noda, T, Tsuge, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
To Be Published
|
|
9JSO
 
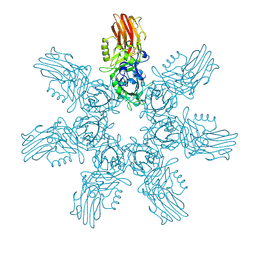 | | Clostridium perfringens iota toxin pore Ib in prepore VIII state | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yamada, T, Sugita, Y, Yoshida, T, Noda, T, Tsuge, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
To Be Published
|
|
9JSM
 
 | | Clostridium perfringens iota toxin pore Ib in prepore VI state | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yamada, T, Sugita, Y, Yoshida, T, Noda, T, Tsuge, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
To Be Published
|
|
9JSL
 
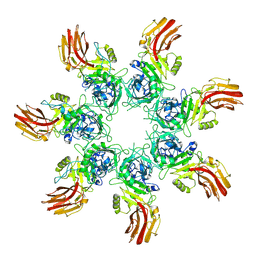 | | Clostridium perfringens iota toxin pore Ib in prepore V state | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yamada, T, Sugita, Y, Yoshida, T, Noda, T, Tsuge, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
To Be Published
|
|
9JSI
 
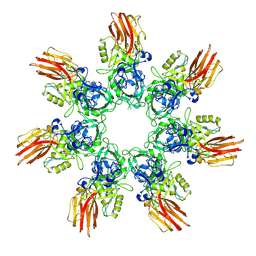 | | Clostridium perfringens iota toxin pore Ib in prepore II state | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yamada, T, Sugita, Y, Yoshida, T, Noda, T, Tsuge, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
To Be Published
|
|
9JSN
 
 | | Clostridium perfringens iota toxin pore Ib in prepore VII state | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yamada, T, Sugita, Y, Yoshida, T, Noda, T, Tsuge, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
To Be Published
|
|
9JSH
 
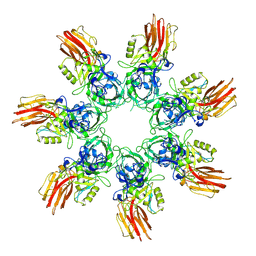 | | Clostridium perfringens iota toxin pore Ib in prepore III state | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yamada, T, Sugita, Y, Yoshida, T, Noda, T, Tsuge, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
To Be Published
|
|
7D7M
 
 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nojima, S, Fujita, Y, Kimura, T.K, Nomura, N, Suno, R, Morimoto, K, Yamamoto, M, Noda, T, Iwata, S, Shigematsu, H, Kobayashi, T. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein.
Structure, 29, 2021
|
|
5Z9W
 
 | | Ebola virus nucleoprotein-RNA complex | | Descriptor: | Ebolavirus nucleoprotein (residues 19-406), RNA (6-MER) | | Authors: | Sugita, Y, Matsunami, H, Kawaoka, Y, Noda, T, Wolf, M. | | Deposit date: | 2018-02-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 angstrom resolution.
Nature, 563, 2018
|
|
6K96
 
 | | Crystal structure of Ari2 | | Descriptor: | Five-membered-cyclitol-phosphate synthase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Miyanaga, A, Tsunoda, T, Kudo, F, Eguchi, T. | | Deposit date: | 2019-06-14 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereochemistry in the Reaction of themyo-Inositol Phosphate Synthase Ortholog Ari2 during Aristeromycin Biosynthesis.
Biochemistry, 58, 2019
|
|
1V94
 
 | | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix | | Descriptor: | isocitrate dehydrogenase | | Authors: | Jeong, J.-J, Sonoda, T, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2004-01-20 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix
Proteins, 55, 2004
|
|
3WC2
 
 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with a tRNA(Phe)(GUG) | | Descriptor: | 76mer-tRNA, Likely histidyl tRNA-specific guanylyltransferase | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.641 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WC0
 
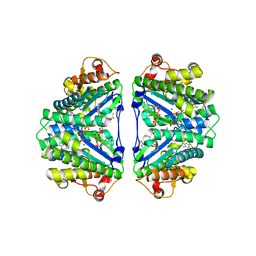 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Likely histidyl tRNA-specific guanylyltransferase, MAGNESIUM ION | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WC1
 
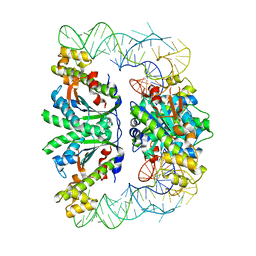 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with a G-1 deleted tRNA(His) | | Descriptor: | 75-mer tRNA, Likely histidyl tRNA-specific guanylyltransferase | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WBZ
 
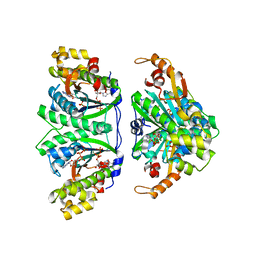 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Likely histidyl tRNA-specific guanylyltransferase, MAGNESIUM ION | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
