2ZMZ
 
 | |
2ZWD
 
 | |
2ZMY
 
 | |
2ZWG
 
 | |
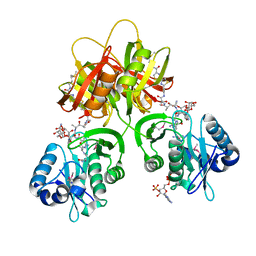
2ZW7
 
 | |
2ZW5
 
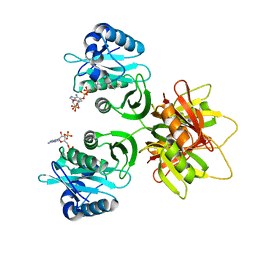 | |
2ZW6
 
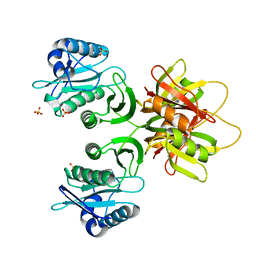 | |
2ZW4
 
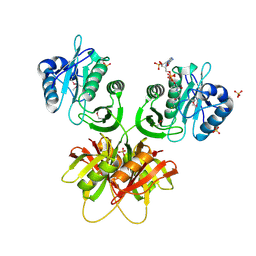 | |
3WSW
 
 | |
3WSV
 
 | |
5DSV
 
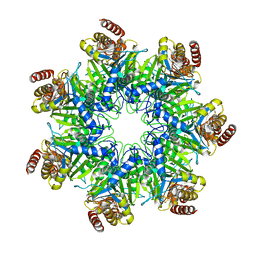 | | Crystal structure of human proteasome alpha7 tetradecamer | | Descriptor: | Proteasome subunit alpha type-3 | | Authors: | Satoh, T, Thammaporn, R, Seetaha, S, Kato, K. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Disassembly of the self-assembled, double-ring structure of proteasome alpha 7 homo-tetradecamer by alpha 6
Sci Rep, 5, 2015
|
|
3WA6
 
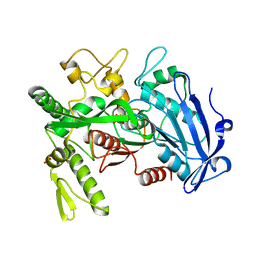 | |
3WA7
 
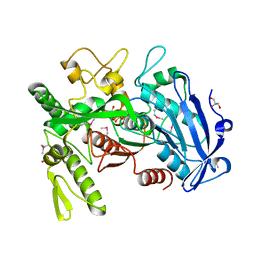 | | Crystal structure of selenomethionine-labeled tannase from Lactobacillus plantarum in the orthorhombic crystal | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Matoba, Y, Tanaka, N, Sugiyama, M. | | Deposit date: | 2013-04-27 | | Release date: | 2013-07-24 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic and mutational analyses of tannase from Lactobacillus plantarum.
Proteins, 81, 2013
|
|
3WCK
 
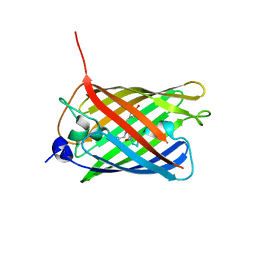 | | Crystal structure of monomeric photosensitizing fluorescent protein, Supernova | | Descriptor: | Monomeric photosenitizing fluorescent protein supernova | | Authors: | Sakai, N, Matsuda, T, Takemoto, K, Nagai, T. | | Deposit date: | 2013-05-27 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation
Sci Rep, 3, 2013
|
|
3AFC
 
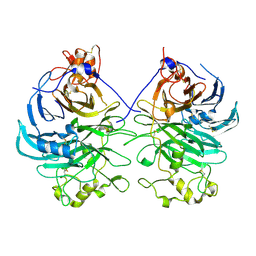 | | Mouse Semaphorin 6A extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin-6A | | Authors: | Yasui, N, Nogi, T, Mihara, E, Takagi, J. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
3AL8
 
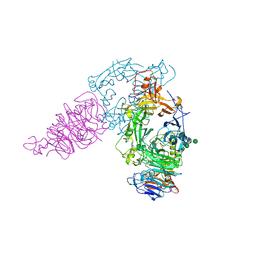 | | Plexin A2 / Semaphorin 6A complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A2, ... | | Authors: | Nogi, T, Yasui, N, Mihara, E, Takagi, J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
3AL9
 
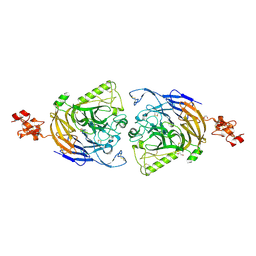 | | Mouse Plexin A2 extracellular domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A2 | | Authors: | Nogi, T, Yasui, N, Mihara, E, Takagi, J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
2ZMX
 
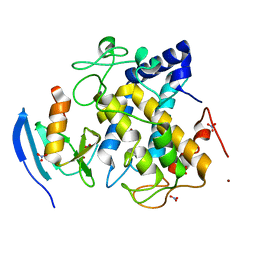 | |
