1MAV
 
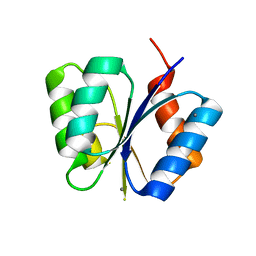 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 6.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1MB3
 
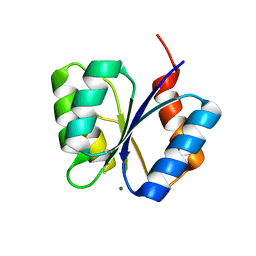 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.5 IN COMPLEX WITH MG2+ | | Descriptor: | MAGNESIUM ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1M5U
 
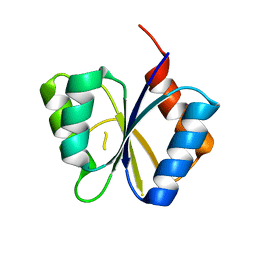 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK. STRUCTURE AT PH 8.0 IN THE APO-FORM | | Descriptor: | cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus
J.Biol.Chem., 277, 2002
|
|
1M5T
 
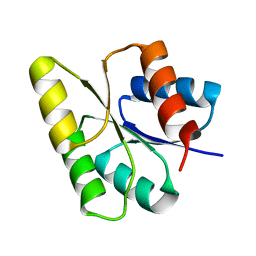 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK | | Descriptor: | cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus
J.Biol.Chem., 277, 2002
|
|
1MB0
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1FV5
 
 | | SOLUTION STRUCTURE OF THE FIRST ZINC FINGER FROM THE DROSOPHILA U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | FIRST ZINC FINGER OF U-SHAPED, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-18 | | Release date: | 2000-10-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1FU9
 
 | | SOLUTION STRUCTURE OF THE NINTH ZINC-FINGER DOMAIN OF THE U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | U-SHAPED TRANSCRIPTIONAL COFACTOR, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1LIQ
 
 | | Non-native Solution Structure of a fragment of the CH1 domain of CBP | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Matthews, J.M, Kwan, A.H.Y, Newton, A, Gell, D.A, Crossley, M, Mackay, J.P. | | Deposit date: | 2002-04-18 | | Release date: | 2002-05-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A New Zinc Binding Fold Underlines the Versatility of Zinc Binding Modules in Protein Evolution
Structure, 10, 2002
|
|
6CVX
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-50 (MK-5172 linear analogue) | | Descriptor: | GLYCEROL, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-4-{[7-methoxy-3-(propan-2-yl)quinoxalin-2-yl]oxy}-L-prolinamide, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
6CVW
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-52 (MK-5172 linear analogue) | | Descriptor: | N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-4-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-L-prolinamide, NS3 protease, SULFATE ION, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
6CVY
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-21 (MK-5172 linear analogue) | | Descriptor: | 3-methyl-N-[(pentyloxy)carbonyl]-L-valyl-(4R)-4-[(3-chloro-7-methoxyquinoxalin-2-yl)oxy]-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-L-prolinamide, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
5VOJ
 
 | |
5VP9
 
 | |
7L7O
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR04-49 | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
7L7L
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-129 | | Descriptor: | 1,1,1-trifluoro-2-methylpropan-2-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
7L7N
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-59 | | Descriptor: | 1,2-ETHANEDIOL, 1-(trifluoromethyl)cyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
7L7P
 
 | |
5ETX
 
 | | Crystal structure of HCV NS3/4A protease A156T variant in complex with 5172-Linear (MK-5172 linear analogue) | | Descriptor: | CHLORIDE ION, NS3 protease, ZINC ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
3SU0
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, Genome polyprotein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.159 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU5
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUE
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUF
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SV7
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU2
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, Genome polyprotein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SV6
 
 | | Crystal structure of NS3/4A protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
