5DNW
 
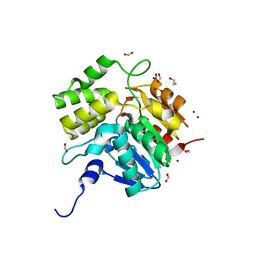 | | Crystal structure of KAI2-like protein from Striga (apo state 1) | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | 登録日 | 2015-09-10 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
5DNU
 
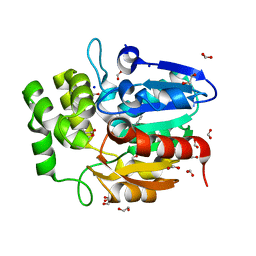 | | Crystal structure of Striga KAI2-like protein in complex with karrikin | | 分子名称: | 1,2-ETHANEDIOL, 3-methyl-2H-furo[2,3-c]pyran-2-one, BENZOIC ACID, ... | | 著者 | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | 登録日 | 2015-09-10 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
5DNV
 
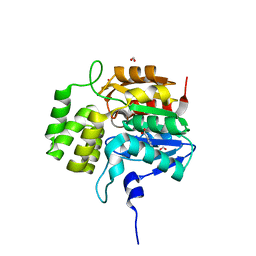 | | Crystal structure of KAI2-like protein from Striga (apo state 2) | | 分子名称: | BENZOIC ACID, FORMIC ACID, ShKAI2iB | | 著者 | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | 登録日 | 2015-09-10 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
1JFC
 
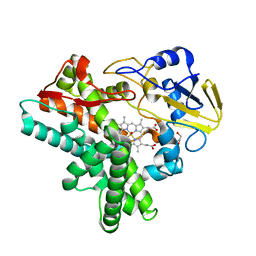 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferrous CO state at atomic resolution | | 分子名称: | CARBON MONOXIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2001-06-20 | | 公開日 | 2001-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8V1L
 
 | |
3HDN
 
 | |
3HDM
 
 | |
1F24
 
 | |
1F25
 
 | |
1F26
 
 | |
1TCH
 
 | |
1TCJ
 
 | |
1TCG
 
 | |
1TCK
 
 | |
1JFB
 
 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferric resting state at atomic resolution | | 分子名称: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, nitric-oxide reductase cytochrome P450 55A1 | | 著者 | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2001-06-20 | | 公開日 | 2001-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HJR
 
 | |
4OI7
 
 | | RAGE recognizes nucleic acids and promotes inflammatory responses to DNA | | 分子名称: | 1,2-ETHANEDIOL, 5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*TP*AP*GP*CP*AP*TP*CP*GP*TP*TP*GP*CP*AP*G)-3', 5'-D(*CP*TP*GP*CP*AP*AP*CP*GP*AP*TP*GP*CP*TP*AP*CP*GP*AP*AP*CP*GP*TP*G)-3', ... | | 著者 | Jin, T, Jiang, J, Xiao, T. | | 登録日 | 2014-01-19 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.104 Å) | | 主引用文献 | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
4OI8
 
 | |
5Z7Y
 
 | | Crystal structure of Striga hermonthica HTL7 (ShHTL7) | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Hyposensitive to light 7, ... | | 著者 | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | 登録日 | 2018-01-30 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z7Z
 
 | | Crystal structure of Striga hermonthica Dwarf14 (ShD14) | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dwarf 14, ... | | 著者 | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | 登録日 | 2018-01-30 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.978 Å) | | 主引用文献 | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z7W
 
 | | Crystal structure of Striga hermonthica HTL1 (ShHTL1) | | 分子名称: | GLYCEROL, Hyposensitive to light 1, MAGNESIUM ION, ... | | 著者 | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | 登録日 | 2018-01-30 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.657 Å) | | 主引用文献 | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z7X
 
 | | Crystal structure of Striga hermonthica HTL4 (ShHTL4) | | 分子名称: | 1,2-ETHANEDIOL, Hyposensitive to light 4, MAGNESIUM ION | | 著者 | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | 登録日 | 2018-01-30 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.055 Å) | | 主引用文献 | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
6J88
 
 | | Crystal structure of HinD with benzo[b]thiophen analog | | 分子名称: | N-[(2S)-1-(1-benzothiophen-3-yl)-3-hydroxypropan-2-yl]-N~2~-methyl-L-valinamide, Nocardicin N-oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Fei, H, Mori, T, Abe, I. | | 登録日 | 2019-01-18 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
6J86
 
 | | Crystal structure of HinD with NMFT | | 分子名称: | N-[(2S)-1-hydroxy-3-(1H-indol-3-yl)propan-2-yl]-Nalpha-methyl-L-phenylalaninamide, Nocardicin N-oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Fei, H, Mori, T, Abe, I. | | 登録日 | 2019-01-18 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
6J85
 
 | | Crystal structure of HinD apo | | 分子名称: | Nocardicin N-oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Fei, H, Mori, T, Abe, I. | | 登録日 | 2019-01-18 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
