3A0Z
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) (nucleotide free form 4: isopropanol, orthorombic) | | Descriptor: | Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A0U
 
 | | Crystal structure of response regulator protein TrrA (TM1360) from Thermotoga maritima in complex with Mg(2+)-BeF (wild type) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A10
 
 | | Crystal structure of response regulator protein TrrA (TM1360) from Thermotoga maritima in complex with Mg(2+)-BeF (SeMet, L89M) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3ETA
 
 | | Kinase domain of insulin receptor complexed with a pyrrolo pyridine inhibitor | | Descriptor: | 1-(3-{5-[4-(aminomethyl)phenyl]-1H-pyrrolo[2,3-b]pyridin-3-yl}phenyl)-3-(2-phenoxyphenyl)urea, insulin receptor, kinase domain | | Authors: | Patnaik, S, Stevens, K, Gerding, R, Deanda, F, Shotwell, B, Tang, J, Hamajima, T, Nakamura, H, Leesnitzer, A, Hassell, A, Shewchuk, L, Kumar, R, Lei, H, Chamberlain, S. | | Deposit date: | 2008-10-07 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3,5-disubstituted-1H-pyrrolo[2,3-b]pyridines as potent inhibitors of the insulin-like growth factor-1 receptor (IGF-1R) tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5XSO
 
 | | Crystal structure of full-length FixJ from B. japonicum crystallized in space group C2221 | | Descriptor: | FORMIC ACID, GLYCEROL, Response regulator FixJ | | Authors: | Nishizono, Y, Hisano, T, Sawai, H, Shiro, Y, Nakamura, H, Wright, G.S.A, Saeki, A, Hikima, T, Yamamoto, M, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system.
Sci Signal, 11, 2018
|
|
5XT2
 
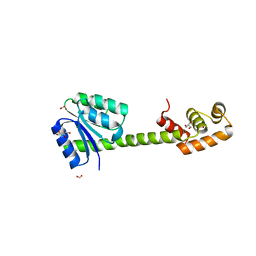 | | Crystal structures of full-length FixJ from B. japonicum crystallized in space group P212121 | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nishizono, Y, Hisano, T, Shiro, Y, Sawai, H, Wright, G.S.A, Saeki, A, Hikima, T, Nakamura, H, Yamamoto, M, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system.
Sci Signal, 11, 2018
|
|
1X3U
 
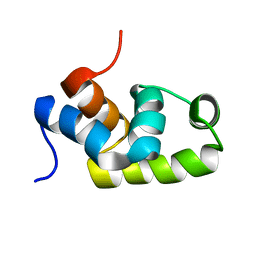 | | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium melilot | | Descriptor: | Transcriptional regulatory protein fixJ | | Authors: | Kurashima-Ito, K, Kasai, Y, Hosono, K, Tamura, K, Oue, S, Isogai, M, Ito, Y, Nakamura, H, Shiro, Y. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium meliloti and its recognition of the fixK promoter
Biochemistry, 44, 2005
|
|
6J82
 
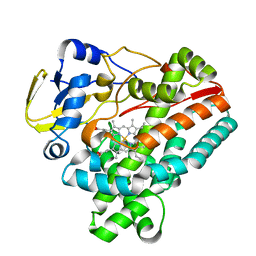 | | Crystal structure of TleB apo | | Descriptor: | Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Alblova, M, Nakamura, H, Mori, T, Abe, I. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
3W0K
 
 | |
3WUQ
 
 | | Structure of the entire stalk region of the dynein motor domain | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1 | | Authors: | Nishikawa, Y, Oyama, T, Kamiya, N, Kon, T, Toyoshima, Y.Y, Nakamura, H, Kurisu, G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the entire stalk region of the Dynein motor domain
J.Mol.Biol., 426, 2014
|
|
5ZHS
 
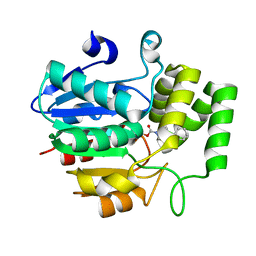 | | Crystal structure of OsD14 in complex with covalently bound KK052 | | Descriptor: | (4-phenylpiperazin-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5ZHT
 
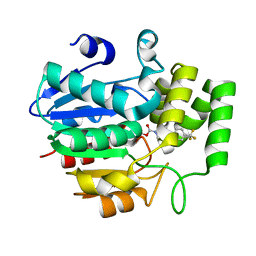 | | Crystal structure of OsD14 in complex with covalently bound KK073 | | Descriptor: | (1H-1,2,3-triazol-1-yl){4-[4-(trifluoromethyl)phenyl]piperazin-1-yl}methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5ZHR
 
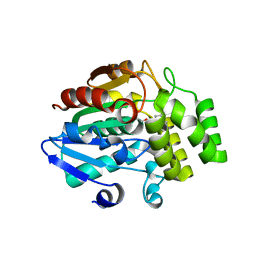 | | Crystal structure of OsD14 in complex with covalently bound KK094 | | Descriptor: | (2,3-dihydro-1H-indol-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
3VXK
 
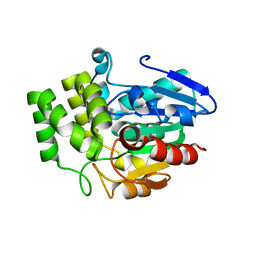 | | Crystal structure of OsD14 | | Descriptor: | Dwarf 88 esterase | | Authors: | Xue, Y.-L, Miyakawa, T, Hou, F, Qin, H.-M, Tanokura, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanism of strigolactone perception by DWARF14
Nat Commun, 4, 2013
|
|
3WIO
 
 | | Crystal structure of OSD14 in complex with hydroxy D-ring | | Descriptor: | (5R)-5-hydroxy-3-methylfuran-2(5H)-one, Probable strigolactone esterase D14 | | Authors: | Xue, Y.-L, Miyakawa, T, Hou, F, Qin, H.-M, Tanokura, M. | | Deposit date: | 2013-09-22 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of strigolactone perception by DWARF14
Nat Commun, 4, 2013
|
|
4FXC
 
 | |
5B57
 
 | | Inward-facing conformation of ABC heme importer BhuUV from Burkholderia cenocepacia | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Hemin import ATP-binding protein HmuV, Putative hemin ABC transport system, ... | | Authors: | Naoe, Y, Nakamura, N, Doi, A, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-04-25 | | Release date: | 2016-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bacterial haem importer complex in the inward-facing conformation
Nat Commun, 7, 2016
|
|
5B58
 
 | | Inward-facing conformation of ABC heme importer BhuUV in complex with periplasmic heme binding protein BhuT from Burkholderia cenocepacia | | Descriptor: | Hemin import ATP-binding protein HmuV, Putative hemin ABC transport system, membrane protein, ... | | Authors: | Naoe, Y, Nakamura, N, Doi, A, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-04-25 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of bacterial haem importer complex in the inward-facing conformation
Nat Commun, 7, 2016
|
|
5DNW
 
 | | Crystal structure of KAI2-like protein from Striga (apo state 1) | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
5DNU
 
 | | Crystal structure of Striga KAI2-like protein in complex with karrikin | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2H-furo[2,3-c]pyran-2-one, BENZOIC ACID, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
5DNV
 
 | | Crystal structure of KAI2-like protein from Striga (apo state 2) | | Descriptor: | BENZOIC ACID, FORMIC ACID, ShKAI2iB | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
2ROM
 
 | |
1F24
 
 | |
1F25
 
 | |
1F26
 
 | |
