4AP4
 
 | | Rnf4 - ubch5a - ubiquitin heterotrimeric complex | | Descriptor: | E3 UBIQUITIN LIGASE RNF4, UBIQUITIN C, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Plechanovova, A, Hay, R.T, Tatham, M.H, Jaffray, E, Naismith, J.H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of a Ring E3 Ligase and Ubiquitin-Loaded E2 Primed for Catalysis
Nature, 489, 2012
|
|
4AKS
 
 | | PatG macrocyclase domain | | Descriptor: | THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AGF
 
 | |
4CQJ
 
 | |
4BG2
 
 | | X-ray Crystal Structure of PatF from Prochloron didemni | | Descriptor: | PATF | | Authors: | Bent, A.F, Koehnke, J, Houssen, W.E, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-03 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of Patf from Prochloron Didemni.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4BQQ
 
 | |
4B5B
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, N-(6-amino-1-benzyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-N,3-dimethylbenzenesulfonamide | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-08-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B4G
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4ARW
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-04-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4AVR
 
 | | Crystal structure of the hypothetical protein Pa4485 from Pseudomonas aeruginosa | | Descriptor: | PA4485 | | Authors: | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Alphey, M.S, Naismith, J.H. | | Deposit date: | 2012-05-29 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B7X
 
 | | Crystal structure of hypothetical protein PA1648 from Pseudomonas aeruginosa. | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROBABLE OXIDOREDUCTASE | | Authors: | Alphey, M.S, McMahon, S.A, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ASJ
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, N-(6-amino-1-benzyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-N-methylbenzenesulfonamide, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B0C
 
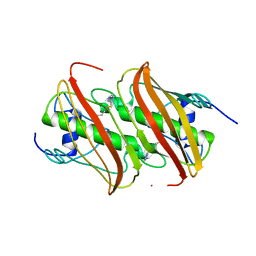 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa in complex with 3-(pentylthio)-4H-1,2,4-triazole | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, 5-(pentylsulfanyl)-1H-1,2,4-triazole | | Authors: | Moynie, L, McMahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2012-07-01 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into the Mechanism and Inhibition of the Beta-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase from Pseudomonasaeruginosa.
J.Mol.Biol., 425, 2013
|
|
4AKT
 
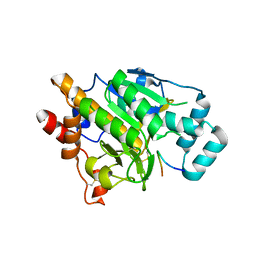 | | PatG macrocyclase in complex with peptide | | Descriptor: | SUBSTRATE ANALOGUE, THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AZS
 
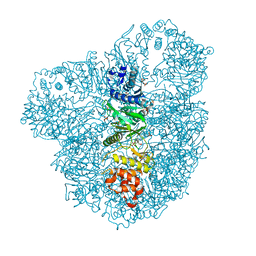 | | High resolution (2.2 A) crystal structure of WbdD. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, METHYLTRANSFERASE WBDD, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
4AZW
 
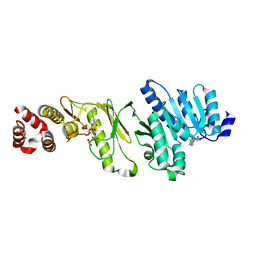 | | Crystal structure of monomeric WbdD. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
4B3U
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-amino-1-benzyl-5-(ethylamino)pyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B0I
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) mutant (H70N) from Pseudomonas aeruginosa in complex with 3-hydroxydecanoyl-N-acetyl cysteamine | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, S-[2-(acetylamino)ethyl] (3R)-3-hydroxydecanethioate | | Authors: | Moynie, L, McMahon, S.A, Leckie, S.M, Duthie, F.G, Koehnke, A, Naismith, J.H. | | Deposit date: | 2012-07-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights Into the Mechanism and Inhibition of the Beta-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase from Pseudomonas Aeruginosa
J.Mol.Biol., 425, 2013
|
|
4B2W
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-18 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B8U
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa in complex with N- isobutyl-2-(5-(2-thienyl)-1,2-oxazol-3-yl-)methoxy)acetamide | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, N-isobutyl-2-{[5-(thiophen-2-yl)-1,2-oxazol-3-yl]methoxy}acetamide, SULFATE ION | | Authors: | Moynie, L, McMahon, S.A, Duthie, F.G, Brenk, R, Naismith, J.H. | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Insights Into the Mechanism and Inhibition of the Beta-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase from Pseudomonasaeruginosa.
J.Mol.Biol., 425, 2013
|
|
4CL6
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa in complex with N-(4- Chlorobenzyl)-3-(2-furyl)-1H-1,2,4-triazol-5-amine | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, N-(4-chlorobenzyl)-5-(furan-2-yl)-1H-1,2,4-triazol-3-amine | | Authors: | Moynie, L, McMahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Substrate Mimic Allows High Throughput Assay of the Faba Protein and Consequently the Identification of a Novel Inhibitor of Pseudomonas Aeruginosa Faba.
J.Mol.Biol., 428, 2016
|
|
4AZT
 
 | | Co-crystal structure of WbdD and kinase inhibitor LY294002. | | Descriptor: | 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, CHLORIDE ION, METHYLTRANSFERASE WBDD, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
4CCV
 
 | | Crystal structure of histidine-rich glycoprotein N2 domain reveals redox activity at an interdomain disulfide bridge: Implications for the regulation of angiogenesis | | Descriptor: | GLUTATHIONE, GLYCEROL, HISTIDINE-RICH GLYCOPROTEIN, ... | | Authors: | McMahon, S.A, Kassaar, O, Stewart, A.J, Naismith, J.H. | | Deposit date: | 2013-10-29 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of Histidine-Rich Glycoprotein N2 Domain Reveals Redox Activity at an Interdomain Disulfide Bridge: Implications for Angiogenic Regulation.
Blood, 123, 2014
|
|
2V7T
 
 | |
2VH3
 
 | | ranasmurfin | | Descriptor: | GLYCEROL, RANASMURFIN, SULFATE ION, ... | | Authors: | Oke, M, Ching, R.T, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, Bloch Junior, C, Botting, C.H, Walsh, M.A, Latiff, A.A, Kennedy, M.W, Cooper, A, Naismith, J.H. | | Deposit date: | 2007-11-17 | | Release date: | 2007-12-04 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Unusual Chromophore and Cross-Links in Ranasmurfin: A Blue Protein from the Foam Nests of a Tropical Frog.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
