7LHT
 
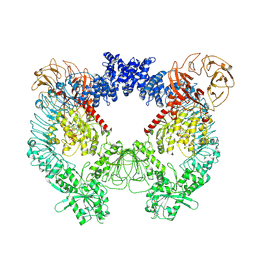 | | Structure of the LRRK2 dimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LHW
 
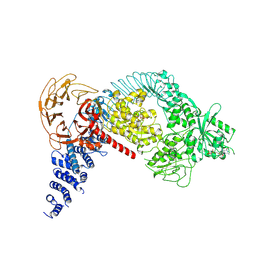 | | Structure of the LRRK2 monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI3
 
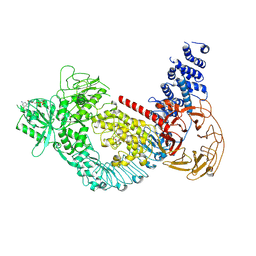 | | Structure of the LRRK2 G2019S mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI4
 
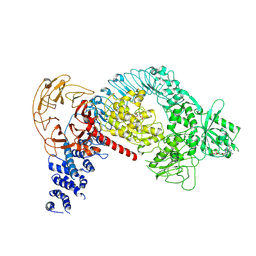 | | Structure of LRRK2 after symmetry expansion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
8AQW
 
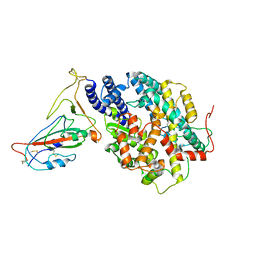 | | BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
9DBQ
 
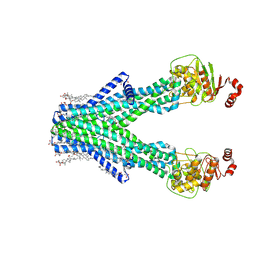 | | Cryo-EM structure of human ABCB6 transporter in an inward-facing conformation | | Descriptor: | ATP-binding cassette sub-family B member 6, CHOLESTEROL HEMISUCCINATE | | Authors: | Shaik, M.M, Myasnikov, A, Oldham, M.L, Baril, S.A, Kalathur, R.C, Schuetz, J.D. | | Deposit date: | 2024-08-23 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The role of ATP-binding Cassette subfamily B member 6 in the inner ear.
Nat Commun, 15, 2024
|
|
8BFN
 
 | | E. coli Wadjet JetABC dimer of dimers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, JetA, JetB, ... | | Authors: | Roisne-Hamelin, F, Beckert, B, Myasnikov, A, Li, Y, Gruber, S. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage.
Mol.Cell, 82, 2022
|
|
6GQ1
 
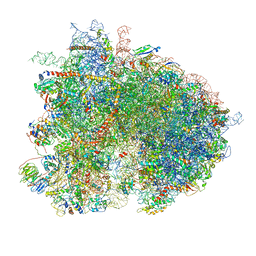 | | Cryo-EM reconstruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GMPPCP/sordarin) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pellegrino, S, Demeshkina, N, Mancera-Martinez, E, Melnikov, S, Simonetti, A, Myasnikov, A, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-11 | | Last modified: | 2018-08-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
7QO7
 
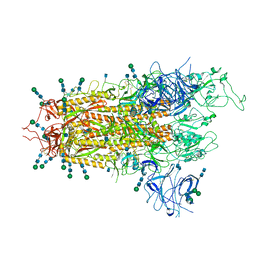 | | SARS-CoV-2 S Omicron Spike B.1.1.529 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Beckert, B, Nazarov, S, Pojer, F, Myasnikov, A, Stahlberg, H, Trono, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion
Biorxiv, 2021
|
|
7QO9
 
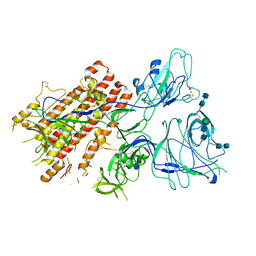 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD and NTD (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,SARS-CoV-2 S Omicron Spike B.1.1.529, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Beckert, B, Nazarov, S, Pojer, F, Myasnikov, A, Stahlberg, H, Trono, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion
Biorxiv, 2021
|
|
8AQU
 
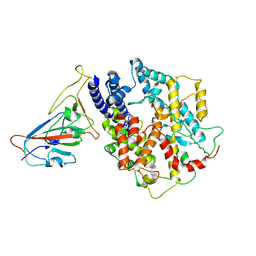 | | BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQV
 
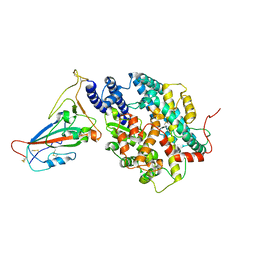 | | BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQS
 
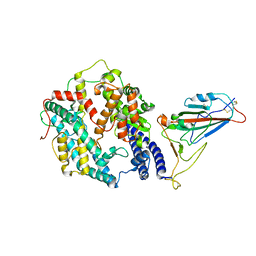 | | BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQT
 
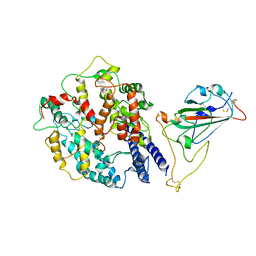 | | Beta SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
5LI0
 
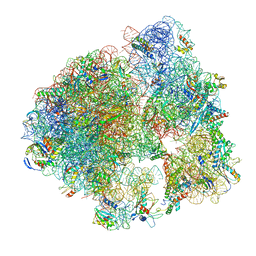 | | 70S ribosome from Staphylococcus aureus | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khusainov, I, Vicens, Q, Bochler, A, Grosse, F, Myasnikov, A, Menetret, J.F, Chicher, J, Marzi, S, Romby, P, Yusupova, G, Yusupov, M, Hashem, Y. | | Deposit date: | 2016-07-13 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the 70S ribosome from human pathogen Staphylococcus aureus.
Nucleic Acids Res., 44, 2016
|
|
8FWK
 
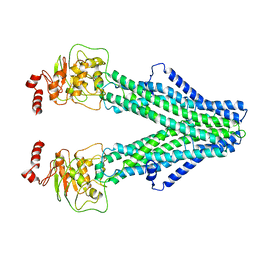 | |
9CPH
 
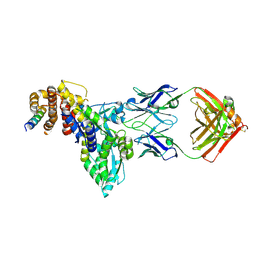 | | Structural basis of BAK sequestration by MCL-1 and consequences for apoptosis initiation | | Descriptor: | Bcl-2 homologous antagonist/killer, Induced myeloid leukemia cell differentiation protein Mcl-1, Synthetic antibody, ... | | Authors: | Uchikawa, E, Myasnikov, A, Dey, R, Moldoveanu, T. | | Deposit date: | 2024-07-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis of BAK sequestration by MCL-1 in apoptosis.
Mol.Cell, 85, 2025
|
|
5ND9
 
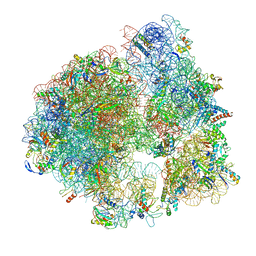 | | Hibernating ribosome from Staphylococcus aureus (Rotated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khusainov, I, Vicens, Q, Ayupov, R, Usachev, K, Myasnikov, A, Simonetti, A, Validov, S, Kieffer, B, Yusupova, G, Yusupov, M, Hashem, Y. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
5ND8
 
 | | Hibernating ribosome from Staphylococcus aureus (Unrotated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khusainov, I, Vicens, Q, Ayupov, R, Usachev, K, Myasnikov, A, Simonetti, A, Validov, S, Kieffer, B, Yusupova, G, Yusupov, M, Hashem, Y. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
6PDY
 
 | | Msp1-substrate complex in open conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PDW
 
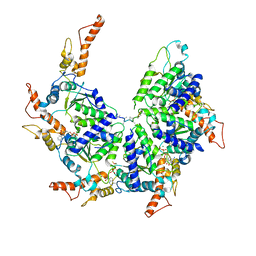 | | Msp1-substrate complex in closed conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PE0
 
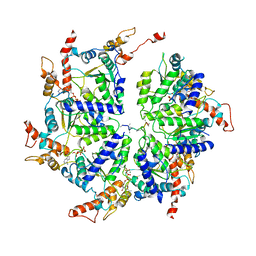 | | Msp1 (E214Q)-substrate complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Membrane-spanning ATPase-like protein, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
8RQB
 
 | | Cryo-EM structure of mouse heavy-chain apoferritin | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Nazarov, S, Myasnikov, A, Stahlberg, H. | | Deposit date: | 2024-01-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (1.09 Å) | | Cite: | Low-dose cryo-electron ptychography of proteins at sub-nanometer resolution.
Nat Commun, 15, 2024
|
|
8EAL
 
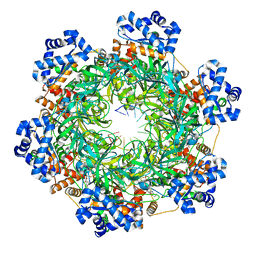 | | SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 1. Merged particles from datasets with 3 different DNA entities | | Descriptor: | DNA, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAI
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 16-mer oligo-dT. Class 2 | | Descriptor: | 16-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
