9ARQ
 
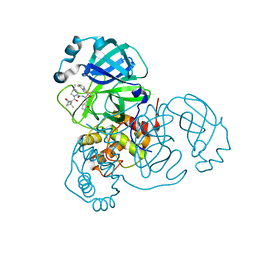 | | Crystal structure of SARS-CoV-2 main protease (authentic protein) in complex with an inhibitor TKB-245 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | 登録日 | 2024-02-23 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ARS
 
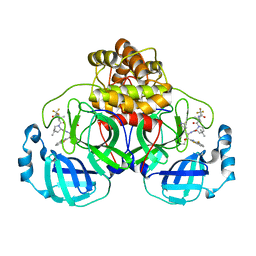 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-245 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | 登録日 | 2024-02-23 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ART
 
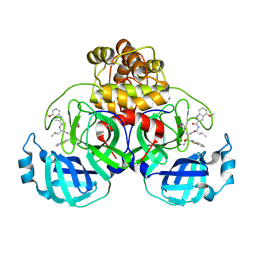 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor 5h | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | 登録日 | 2024-02-23 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | 登録日 | 2024-03-04 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
3P0B
 
 | |
2OM7
 
 | | Structural Basis for Interaction of the Ribosome with the Switch Regions of GTP-bound Elongation Factors | | 分子名称: | 16S ribosomal RNA (H5), 30S ribosomal protein S12, 30S ribosomal protein S2, ... | | 著者 | Connell, S.R, Wilson, D.N, Rost, M, Schueler, M, Giesebrecht, J, Dabrowski, M, Mielke, T, Fucini, P, Spahn, C.M.T. | | 登録日 | 2007-01-21 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors.
Mol.Cell, 25, 2007
|
|
1IUH
 
 | | Crystal structure of TT0787 of thermus thermophilus HB8 | | 分子名称: | 2'-5' RNA Ligase | | 著者 | Kato, M, Sakai, H, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-03-05 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the 2'-5' RNA Ligase from Thermus thermophilus HB8
J.MOL.BIOL., 329, 2003
|
|
7X5O
 
 | | Crystal structure of E. faecium SHMT in complex with Me-THF and PLP-Gly | | 分子名称: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Serine hydroxymethyltransferase | | 著者 | Hasegawa, K, Hayashi, H. | | 登録日 | 2022-03-05 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Serine hydroxymethyltransferase as a potential target of antibacterial agents acting synergistically with one-carbon metabolism-related inhibitors.
Commun Biol, 5, 2022
|
|
7X5N
 
 | | Crystal structure of E. faecium SHMT in complex with (+)-SHIN-1 and PLP-Ser | | 分子名称: | (4R)-6-azanyl-4-[3-(hydroxymethyl)-5-phenyl-phenyl]-3-methyl-4-propan-2-yl-1H-pyrano[2,3-c]pyrazole-5-carbonitrile, Serine hydroxymethyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | 著者 | Hasegawa, K, Hayashi, H. | | 登録日 | 2022-03-05 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Serine hydroxymethyltransferase as a potential target of antibacterial agents acting synergistically with one-carbon metabolism-related inhibitors.
Commun Biol, 5, 2022
|
|
7VGU
 
 | | Time-resolved serial femtosecond crystallography structure of light-driven chloride ion-pumping rhodopsin, NM-R3 : structure obtained 1 msec after photoexcitation with bromide ion | | 分子名称: | BROMIDE ION, Chloride pumping rhodopsin, DECANE, ... | | 著者 | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | 登録日 | 2021-09-18 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VGT
 
 | | Time-resolved serial femtosecond crystallography structure of light-driven chloride ion-pumping rhodopsin, NM-R3: resting state structure with bromide ion | | 分子名称: | BROMIDE ION, Chloride pumping rhodopsin, DECANE, ... | | 著者 | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | 登録日 | 2021-09-18 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VGV
 
 | | Anion free form of light-driven chloride ion-pumping rhodopsin, NM-R3, structure determined by serial femtosecond crystallography at SACLA | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, HEXADECANE, ... | | 著者 | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | 登録日 | 2021-09-18 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7BPG
 
 | | Structure of serinol nucleic acid - RNA complex | | 分子名称: | CALCIUM ION, RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3'), SNA (S-(F7R)(F7X)(F7O)(F7R)(F7X)(F7O)(F7R)(F7U)-R) | | 著者 | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | 登録日 | 2020-03-22 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
7BPF
 
 | | Structure of L-threoninol nucleic acid - RNA complex | | 分子名称: | L-aTNA (3'-(*GP*CP*AP*GP*CP*AP*GP*C)-1'), RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3') | | 著者 | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | 登録日 | 2020-03-22 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
1WNG
 
 | |
1WJX
 
 | | Crystal sturucture of TT0801 from Thermus thermophilus | | 分子名称: | POTASSIUM ION, SsrA-binding protein | | 著者 | Bessho, Y, Shibata, R, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-29 | | 公開日 | 2004-11-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2YW8
 
 | | Crystal structure of human RUN and FYVE domain-containing protein | | 分子名称: | RUN and FYVE domain-containing protein 1, SULFATE ION, ZINC ION | | 著者 | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-20 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of human RUN and FYVE domain-containing protein
To be Published
|
|
1WV9
 
 | | Crystal Structure of Rhodanese Homolog TT1651 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 | | 分子名称: | Rhodanese Homolog TT1651 | | 著者 | Mizohata, E, Hattori, M, Tatsuguchi, A, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-12-12 | | 公開日 | 2005-06-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the single-domain rhodanese homologue TTHA0613 from Thermus thermophilus HB8
Proteins, 64, 2006
|
|
1WV8
 
 | | Crystal structure of hypothetical protein TTHA1013 from an extremely thermophilic bacterium thermus thermophilus HB8 | | 分子名称: | hypothetical protein TTHA1013 | | 著者 | Mizohata, E, Hattori, M, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-12-12 | | 公開日 | 2005-06-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the hypothetical protein TTHA1013 from Thermus thermophilus HB8
Proteins, 61, 2005
|
|
2YY0
 
 | | Crystal Structure of MS0802, c-Myc-1 binding protein domain from Homo sapiens | | 分子名称: | C-Myc-binding protein | | 著者 | Xie, Y, Wang, H, Ihsanawati, K.T, Kishishita, S, Takemoto, C, Shirozu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-27 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | crystal structure of c-Myc-1 binding protein domain from Homo sapiens
To be Published
|
|
2CYC
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Pyrococcus horikoshii | | 分子名称: | TYROSINE, tyrosyl-tRNA synthetase | | 著者 | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Sakamoto, K, Terada, T, Shirouzu, M, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-06 | | 公開日 | 2005-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
2CYB
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Archaeoglobus fulgidus | | 分子名称: | TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-06 | | 公開日 | 2005-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
2YVR
 
 | | Crystal structure of MS1043 | | 分子名称: | Transcription intermediary factor 1-beta, ZINC ION | | 著者 | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-13 | | 公開日 | 2008-04-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of MS1043
To be Published
|
|
2YV8
 
 | | Crystal structure of N-terminal domain of human galectin-8 | | 分子名称: | Galectin-8 variant | | 著者 | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-10 | | 公開日 | 2008-04-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal structure of N-terminal domain of human galectin-8
To be Published
|
|
2E0T
 
 | | Crystal structure of catalytic domain of dual specificity phosphatase 26, MS0830 from Homo sapiens | | 分子名称: | Dual specificity phosphatase 26 | | 著者 | Xie, Y, Kishishita, S, Murayama, K, Hori-Takemoto, C, Chen, L, Liu, Z.J, Wang, B.C, Shirozu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-10-13 | | 公開日 | 2007-10-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | High-resolution crystal structure of the catalytic domain of human dual-specificity phosphatase 26.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
